Short Communication
DNA Barcoding: Amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species
Javed Iqbal Wattoo*1†, Muhammad Zafar Saleem2†, Muhammad Saqib Shahzad1, Amina Arif1, Amir Hameed3, Mushtaq Ahmad Saleem1
Adv. life sci., vol. 4, no. 1, pp. 03-07, November 2016
*- Corresponding Author: Javed Iqbal Wattoo (Email: dr.javed@ucp.edu.pk)
Authors' Affiliations
2- Centre for Applied Molecular Biology (CAMB), University of Punjab, Lahore – Pakistan
3- Department of Bioinformatics and Biotechnology, Government College University, Faisalabad – Pakistan
† Equal Contributors
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: DNA barcoding is a novel method of species identification based on nucleotide diversity of conserved sequences. The establishment and refining of plant DNA barcoding systems is more challenging due to high genetic diversity among different species. Therefore, targeting the conserved nuclear transcribed regions would be more reliable for plant scientists to reveal genetic diversity, species discrimination and phylogeny.
Methods: In this study, we amplified and sequenced the chloroplast DNA regions (matk+rbcl) of Solanum nigrum, Euphorbia helioscopia and Dalbergia sissoo to study the functional annotation, homology modeling and sequence analysis to allow a more efficient utilization of these sequences among different plant species. These three species represent three families; Solanaceae, Euphorbiaceae and Fabaceae respectively. Biological sequence homology and divergence of amplified sequences was studied using Basic Local Alignment Tool (BLAST).
Results: Both primers (matk+rbcl) showed good amplification in three species. The sequenced regions reveled conserved genome information for future identification of different medicinal plants belonging to these species. The amplified conserved barcodes revealed different levels of biological homology after sequence analysis. The results clearly showed that the use of these conserved DNA sequences as barcode primers would be an accurate way for species identification and discrimination.
Conclusion: The amplification and sequencing of conserved genome regions identified a novel sequence of matK in native species of Solanum nigrum. The findings of the study would be applicable in medicinal industry to establish DNA based identification of different medicinal plant species to monitor adulteration.
Key words: DNA Barcoding, Medicinal Plants, matk, rbcl, BLAST, Species Identification
Introduction
DNA barcoding relies on finding different conserved regions in divergent species to produce a large scale reference genome library. DNA barcoding is universally accepted and reliable method for species identification. The nuclear and mitochondrial sequences are mapped and sequenced in different species to design DNA based barcode primers. These barcodes are used for phylogenetic analysis, genetic diversity and species discrimination in different organisms. This system not only helps to classify the organisms but also reveals genetic information for species ancestral inheritance and flagging of new species [1,2]. In plants, finding effective and robust conserved regions are more challenging because of high genome diversity. Chloroplasts are active metabolic machinery in green plants to convert light energy to carbohydrates. High-throughput sequencing technology has resulted sequencing of about 800 chloroplast genomes from different plants [3]. Two conserved regions from plastid (chloroplast) genome (matk+rbcl) were proposed as barcode primers to discriminate large group of angiosperms.
About 80% of world population rely on herbal medicine for some aspects of their primary health care [4]. A drug could be lethal if it is adulterated with toxic materials. Molecular barcoding is the preferred method to monitor the adulteration in medicinal plants. Solanum nigrum and Euphorbia helioscopia are well known medicinal plant species for their therapeutic properties, and have been extensively used as antioxidant, antitumorigenic [5,6], hepatoprotective [7,8], anti-inflammatory [9], diuretic, and antipyretic [10]. For successful barcoding experiment, sequence difference between two species must be higher enough to discriminate both. However, in case of intra and interspecific variation, the sequence variation must be lower enough to distinguish the variations. Seven chloroplast genomic regions were evaluated by Consortium for the Barcode of Life Plant Working Group and finally, a combination of matK+rbcl was suggested as universal barcode for identification and authentication of flowering plants [11]. The universal primer rbcl has high universality but low resolution whereas matK offers low universality but high resolution among different species. A combination of both markers matK+rbcl can help to discriminate maximum number of species. However, in order to attain maximum discrimination rate between two closely related species, a combination of Internal Transcribed Spacer (ITS+matK+rbcl) was suggested by China Plant BOL Group [12]. DNA is more stable macromolecule than RNA and is found in all plant tissues. Therefore, DNA based markers are preferred for precise identification of medicinal plants.
The aims of current study were four fold: (1) amplification and sequencing of matK and rbcl regions in three plant species (2) to study functional annotation and homology modelling of both sequences (matk+rbcl) among three species using Basic Local Alignment Search Tool (BLAST), (3) comparative genomic analysis in order to study these sequences in detail among different medicinal plants (4) to explore the relationship between Solanum nigrum, Euphorbia helioscopia and Euphorbia helioscopia. The progress in the study would further be helpful for identification, authentication and to monitor the adulteration of medicinal biomaterials under national and international trade.
Methods
Plant Material
Three plant species (Solanum nigrum, Euphorbia helioscopia and Dalbergia sissoo) were used in the study. The former plant species have significant role in herbal medicine industry and have been extensively used in various traditional systems of medicines worldwide. The later plant species (Dalbergia sissoo) is the tree of economic importance in forest based industry. All the three species were grown and maintained at Centre for Applied Molecular Biology (CAMB), Lahore, Pakistan under controlled conditions.
DNA Extraction
Young leaf samples were collected from each plant and crushed in liquid nitrogen to get fine powder using sterile mortar and pestle. DNA was extracted using DNeasy mini kit Qiagen (Cat # 74104). The quality of extracted DNA was further estimated using nanodrop method (Thermo- scientific, Wilmington, USA). The isolated DNA was stored at -20°C.
Amplification
A PCR reaction mixture of 50 µL comprised the following: 3 µL of template DNA (300 ng), 5 µL PCR buffer (10X), 5 µL MgCl2 (25mM), 5 µL of both forward and reverse primers (10 pmol) (Table 1), 0.35 µL Taq polymerase, 5 µL dNTPs (1mM) and the rest was added with deionized distilled water. PCR based amplification of barcoding regions was performed using 96-well thermal cycler (Applied Biosystems) and the profile was adjusted to 35 cycles as follows: 94°C for 30 seconds, followed by annealing at 54°C for 45 seconds, and then by extension at 72°C for 45 seconds. After 35 cycles, the profile was linked on hold at 4°C. The amplified PCR products were electrophoresed using 1% agarose gel (1x TAE buffer & 0.5 μg/mL ethidium bromide) to check the presence or absence of bands. Gel imaging was done using UVP gel imaging system. The band size of amplified products was determined using 100 bp ladder (GE Healthcare).
Nucleotide Sequencing
The PCR amplified products were excised from 1% TAE gel and purified using DNA purification kit (Thermo scientific). The purified samples were again quantified with UV-2800 spectrophotometer (BMS). The sequencing reaction of 20 µL was prepared as follows: 2 µL of template DNA (20 ng), 1 µL sequencing buffer (5X) and 2 µL big dye terminator (ABI, version 3.1). The final volume was adjusted to 20 µL with sterile distilled water. The thermal profile was adjusted to 35 cycles as follows: 95°C for 5 minutes, 94°C 1 minute, 53°C 45 seconds, 68°C 4 minutes and final extension was done at 68°C for 10 minutes. Sequenced products were precipitated using 2 volume of 80% propanol and then washed twice with 80% ethanol. The products were air dried and re-suspended in 15 µL formamide denaturing buffer. The final sequencing was done using ABI DNA sequencer following standard protocol. Each sample was done in triplicate.
Results
PCR based amplification of conserved regions (matk and rbcl) is primarily required to establish DNA barcodes for species identification. With both universal primers rbcl and matk, we observed good results of PCR amplification in three species (Solanum nigrum, Euphorbia helioscopia & Euphorbia helioscopia). Both primers resulted in efficient amplification (Figure 1). The results validate the previous findings of using seven plastid regions (rbcL, matK, rpoC1, rpoB, trnH-psbA, atpF-atpH & psbK-psbI) in angiosperms, gymnosperms and liverwort [13].
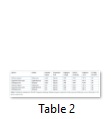
Sequence homology of the amplified sequences was detected using Basic Local Alignment Tool (BLAST). The sequence length of matk was 549, 852 and 296 nucleotides with Solanum nigrum, Euphorbia helioscopia and Dalbergia sissoo respectively (Table 2). Similarly with rbcl, the sequence length of three species was 632, 620 and 572 nucleotides respectively (Table 1). With matk, the sequence homology of Solanum nigrum and Euphorbia helioscopia was 94% and 95% respectively while Dalbergia sissoo showed 0% sequence homology (Table 2). Similarly with rbcl, sequence homology of all the three species was 96%, 99% and 96% respectively (Table 2).
Tables & Figures
Discussion
The genome sequence analysis of many species followed by mapping of complex traits associated with divergent phenotypes resulted in identification of many genes and their detailed inheritance patterns in many plant species. In PCR based amplification, quality of DNA plays an important role. Sometimes, the presence of metabolites in medicinal plants effect DNA quality during isolation and even closely related species may require different DNA isolation protocols [14]. The sequence variation from reference sequence and phylogenetic reconstruction is the basic principle for species identification in plants [15]. The use of DNA based markers (except RFLP) as universal primers has significant results in species identification as they result in good amplification across different genomic regions among divergent species [16]. Next generation sequencing is another facility of advanced genomics era to have more precise picture of species genome and to identify more orthologous and paralogous regions at different loci of different species. Sequencing of entire plastid genome along with ITS & nrDNA is becoming method of choice for species discriminations [17]. In the current study, we amplified and sequence the conserved regions in three different plant species. These species represent huge proportions of medicinal samples in pharmaceutical industry. These DNA barcodes were first time applied in native species which would be a pioneer step to establish DNA based monitoring protocols for medicinal adulteration under national and international trade. However, there are still lot of plant genomes lacking sequence information. A genus-based identification system would be a method of choice in species identification in indigenous land plants. Re-sequencing of additional loci for target based improvements would relatively be helpful to detect more conserved genomic regions among different plant species.
The findings of current study suggest that the use of universal primers (rbcL+matK) for DNA barcoding is successful for amplification, identification and discrimination of above mentioned indigenous plant species. The amplification success rates were 100% for all the three species. Both the primer pairs resulted sharp bands that were required for reliable DNA sequencing. Thus, using DNA barcodes primers systems is reliable, fast and cheap system for identification of medicinal plants at genus and species level of indigenous land species. The recommended pair combination of rbcL+matK discriminate about 90% of flowering plants (CBOL 5), however, more conserved genomic regions could be identified and used to detect adulteration of medicinal material. Generation of extensive DNA data base (using next generation sequencing) while focusing more conserved regions would be powerful for medicinal plants identification [18,19]. These records would further be helpful to study taxonomy, ecology, phylogeny and morphology of different species [20]. However, the development of new protocols and amplification strategies with new primer cocktails would significantly revolutionize the field of DNA barcoding by depicting more detailed genome information from different species.
References
- Son YO, Kim J, Lim JC, Chung Y, Chung GH, et al. Ripe fruits of Solanum nigrum L. inhibits cell growth and induces apoptosis in MCF-7 cells. Food and Chemical Toxicology, (2003); 41(10): 1421-1428.
- Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences USA, (2004); 101(41): 14812-14817.
- Kelchner SA. The evolution of non-coding chloroplast DNA and its application in plant systematics. Annals of the Missouri Botanical Garden, (2000); 87(4): 482-498.
- Hebert PD, Cywinska A, Ball SL. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, (2003); 270(1512): 313-321.
- Glossman MD. CHIH-DFT determination of the molecular structure and infrared and ultraviolet spectra of-solanine. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, (2007); 66(1): 208-211.
- Sikdar M, Dutta U. Traditional phytotherapy among the Nath people of Assam. Ethno-med, (2008); 2(1): 39-45.
- Sharma Y, Singh H, Mehra B. Hepatoprotective effect of few Ayurvedic herbs in patients receiving antituberculus treatment. Indian Journal of Traditional Knowledge, (2004); 3391-396.
- Kaushik D, Jogpal V, Kaushik P, Lal S, Saneja A, et al. Evaluation of activities of Solanum nigrum fruit extract. Archives of Applied Science Research, (2009); 1(1): 43-50.
- Ravi V, Saleem TM, Patel S, Raamamurthy J, Gauthaman K. Anti-inflammatory effect of methanolic extract of Solanum nigrum Linn berries. International Journal of Applied Research in Natural Products, (2009); 2(2): 33-36.
- GonzálezTE, Tapia E, López‐Meraz L, Navarrete A, Reyes‐Ramírez A, et al. Anticonvulsant Effect of Annona diversifolia Saff. and Palmitone on Penicillin‐induced Convulsive Activity. A Behavioral and EEG Study in Rats. Epilepsia, (2006); 47(11): 1810-1817.
- Jinbo U, Kato T, Ito M. Current progress in DNA barcoding and future implications for entomology. Entomological Science, (2011); 14(2): 107-124.
- Li FW, Kuo LY, Rothfels CJ, Ebihara A, Chiou WL, et al. rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS One, (2011); 6(10): e26597.
- Group CPW, Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, et al. A DNA barcode for land plants. Proceedings of the National Academy of Sciences USA, (2009); 106(31): 12794-12797.
- Khanuja SP, Shasany AK, Darokar MP, Kumar S. Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Molecular Biology Reporter, (1999); 17(1): 74-74.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, (1997); 25(17): 3389-3402.
- Kumar P, Gupta V, Misra A, Modi D, Pandey B. Potential of molecular markers in plant biotechnology. Plant Omics, (2009); 2(4): 141.
- Waugh J. DNA barcoding in animal species: progress, potential and pitfalls. BioEssays, (2007); 29(2): 188-197.
- Padial JM, Miralles A, De la Riva I, Vences M. The integrative future of taxonomy. Frontiers in Zoology, (2010); 7(1): 1.
- Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature, (2005); 435(7038): 17-17.
- Hajibabaei M, Singer GA, Hebert PD, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends in Genetics, (2007); 23(4): 167-172.