Full Length Research Article
Marker Assisted Selection for Relative Water Content, Excised leaf Water Loss and Cell Membrane Stability in Cotton
Muhammad Asif Saleem*1, Muhammad Waqas Amjid2, Muhammad Qadir Ahmad1, Etrat Noor1, Abdul Qayyum1, Masood Iqbal Awan3, Muhammad Asif1, Muhammad Nauman1
Adv. life sci., vol. 5, no. 2, pp. 56-60, February 2018
*- Corresponding Author: Muhammad Asif Saleem (Email: asifsaleempbg@gmail.com)
Authors' Affiliations
2- Department of Plant Breeding and Genetics, PMA-Arid University, RWP
3- Department of Agronomy, University of Agriculture Faisalabad
Abstract![]()
Introduction
Methods
Results
Discussion
References
Supplementary Data
Abstract
Background: Drought stress is a major limitation in agricultural productivity. In cotton, drought tolerance is a multi-genic trait. The quantitative trait loci (QTLs), conferring drought tolerance in cotton, could be exploited for stress breeding using marker assisted selection.
Methods: We have screened drought related varieties of Pakistan using DNA markers to identify reported QTLs for drought tolerance. A total of 44 of these varieties were selected. All varieties were sown in the field to record relative water content, excised leaf water loss and cell membrane stability under drought stress condition. QTLs for relative water content, excised leaf water loss and cell membrane stability were checked from all varieties by using DNA markers NAU-2954, NAU-2715, NAU-6672, NAU-8406 and NAU-6790.
Results: Genotypic and phenotypic results showed that the QTL for relative water content qtlRWC-1 present on chromosome 23, linked marker NAU-2954, could be a major QTL conferring drought tolerance in cotton. Using Marker Assisted Selection the variety CRIS-134 showed all concerned QTLs for drought tolerance.
Conclusion: QTL for relative water content qtlRWC-1 could be a major QTL for drought stress tolerance in cotton. The variety CRIS-134 may be used for breeding drought tolerant cultivar.
Key words: Cotton, DNA Marker, Drought Stress activity, Phytotoxicity
Introduction
For agricultural crops, availability of fresh water is declining day by day. Water contents in plants are decreased as soil moisture level decreases [1]. As stomatal conductance became low, CO2 uptake decreased [2]. Under drought stress, ATP content decreases as the drought is increased [3]. Under drought stress, normal functioning in plants is disturbed [4]. As compared with other crops, drought stress drastically affects cotton, the most important fiber crop of the world [5-9]. In cotton, drought stress lowers leaf water content and cell membrane stability [10-12], cellular growth [13,14], roots and stem growth [15], number of bolls per plant [16] and ultimately reduces yield [17,18].
Drought stress tolerance in cotton is a complex quantitative trait. The traits contributing drought tolerance are morphological as well as physiological such as relative water content, excised leaf water loss and cell membrane stability. The mechanism in plant which helps maintain water content in leaves during drought stress could enhance drought tolerance [19,20]. The most important defense trait under drought stress is relative water content, used as measurement of water retention capacity in plants [21,22]. Similarly, another defense mechanism under drought stress is to maintain cell membrane integrity [23]. The exploitation of genetic control of these traits could help enhance breeding for drought tolerance.
In crop plants, QTLs mapping for many important traits, such as yield, quality, disease resistance and drought tolerance is an ongoing research objective for researchers around the world. The identification of genomic regions carrying genes associated with quantitative trait called quantitative trait loci. DNA markers linked to the QTLs could be used for selection or screening of germplasm to find QTLs for the traits of interest. In cotton, QTLs mapping has been conducted for various traits such as fiber traits [24,25], boll traits [26], chlorophyll contents [27-29], earliness [30-32] and yield traits [33-36].
QTLs for the traits conferring drought tolerance such as relative water content [37,38], excised leaf water loss [38], cell membrane stability [37], osmotic potential and osmotic adjustment [39] has been identified. DNA markers linked with these traits could be used for marker assisted selection for drought tolerance in cotton.
Using DNA markers for various important traits, selection/screening/validation has been conducted for many traits such as disease resistance in tomato [40], fragrance genes in rice [41] and in wheat for rust resistance [42]. Very little attention has been given to screen germplasm for drought tolerance using DNA markers along with phenotypic screening. This research work aims to use marker assisted selection to screen out variety/genotype containing QTLs for drought tolerance.
Methods
All 44 varieties (Table 2) were selected on the basis of relevancy with drought tolerance. The collected varieties were evaluated using RCBD in three replications. In each replication, there were two rows for each variety. Each row comprised of ten plants, planted at 30cm plant to plant distance and 75 cm row to row distance. Irrigation was applied once after 40 days of planting. Physiological data related to drought tolerance was recorded when plants, under drought stress, showed effects of drought stress.
Relative Water Content (RWC)
Leave samples from plant were taken for analysis. After fresh weight, all samples were dipped in water to measure turgid weight. Finally the samples were oven dried (70oC) to measure dry weight. The RWC was calculated using formula as by [43].
RWC = [(Fresh weight-Dry weight) / (Turgid weight-Dry weight)] ×100
Excised Leaf Water Loss (ELWL)
Three leaves per plant were taken. Using electric balance, fresh weight was measured, followed by keeping leaf samples on bench at normal room temperature. Wilted weight was recorded after twenty four hours. Finally samples were oven dried (at 70°C) to record dry weight. Using formula given by [44] ELWL was calculated
ELWL = [(Fresh weight-Wilted weight)/Dry weight]
Cell Membrane Stability (CMS)
From each plant, three leave samples were used to measure CMS using following formula by [45]:
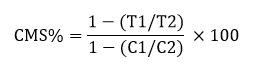
Where, T1= Stress sample conductance before autoclaving.
T2= Stress sample conductance after autoclaving
C1= Control sample conductance before autoclaving
C2= Control sample conductance after autoclaving
DNA Marker Studies
The leaves of all 44 varieties were used for DNA extraction. The selected leaves were frozen at -80oC. DNA was extracted by using standard CTAB method [46]. The DNA samples were checked for quality by gel electrophoresis. The DNA samples giving smear in the gel were rejected and only good quality DNA was selected for PCR studies. All 44 varieties were screened with DNA markers (Table 1). These markers were selected for relative water content, excised leaf water loss and cell membrane stability. PCR products were run on 1% agarose gel. Results were saved through Gel Documentation.
Results
The analysis of variance showed that all 44 varieties showed significant variation for the traits relative water content, excised leaf water loss and cell membrane stability (Table 2 in supplementary data). The data for physiological traits is given in Table 1. DNA marker studies shows that among 44 selected drought tolerant varieties, few varieties showed containing QTLs for drought tolerance. DNA marker 2954, band size 150bp, linked with qtlRWC-1 and qtlELWL at chromosome no 23 [38] was appeared in variety 124-F, BH-36, CIM-448, NIAB-999, CIM-707, 149-F, MNH-93, CRIS-134, 199-F, CIM-506, MS-39, CIM-482, LSS, NIAB-78, MNH-552, CIM-109, BH-118, B-557, 268-F, MNH-554, 238-F, 4F and CIM-70 (Fig. 1) depicts presence of both QTLs. The size of band was similar as reported for these QTLs [38]. Phenotypic data (Table 2) also shows high relative water content with an average of 70% and low excised leaf water loss with an average of 2.1 in these genotypes, showing tolerance against drought stress.
DNA marker NAU-2715 linked with qtlRWC-2 [38] was present in NIAB-111, AC-134, MNH-147, S-12, CIM-707, BH-160, 149-F, MNH-93, CRIS-134, 199-F, S-14, MS-39, CRIS-9, K-68/9, MNH-129, B-557, MS-40, 216-F and 216-F (Fig. 1). Phenotypic data (Table 2) shows moderate water percentage with an average of 55%. DNA marker NAU-6672 linked with qtlRLWC1 [37] was appeared only in MNH-147, MNH-93 and CRIS-134. DNA marker NAU-8406 linked with qtlELWL [37] was detected only in NIAB-111, MNH-552 and CRIS-134 with moderate values of excised leaf water loss (Table 2 in supplementary data). Genotypic and phenotypic data shows that this QTL may not be considered as major contributor for the trait.
Tables & Figures
Discussion
Screening with DNA markers and recording of phenotypic data shows that the DNA marker NAU-2954 linked with two QTLs, one for relative water content qtlRWC-1 and one for excised leaf water loss qtlELWL contribute high drought tolerance in cotton. This QTL was reported to be mapped on chromosome no 23 at LOD=2.74 [38]. The additive effect for this QTL was 4.87, showing high inheritance. The varieties also showed high drought tolerance for relative water content and excised leaf water loss (Table 1). High drought tolerance was associated with presence of these QTLs shows that these QTLs may be considered as major contributors for drought tolerance in cotton. To breed drought tolerant cultivars, the genotypes carrying these QTLs may be included in the breeding programs.
Although a numbers QTLs has been detected for many important traits in many crops but less efforts has been made for examination of these QTLs in germplasm. In this research work some important QTLs has been screened in cotton varieties, considered as drought tolerant in Pakistan. Among these some varieties showed high relative water content as well as showed presence of QTL relevant to the trait. Similarly some varieties showed low excised leaf water loss as well as QTL for the trait. Among these, some varieties showed high relative water content and low excised leaf water loss as well as two QTLs namely qtlRWC-1 and qtlELWL like CRIS-134. The data represents the high value of these QTLs for drought tolerance in cotton. A variety MNH-554 was tolerant in physiological screening but with absence of QTLs under study, depicts the need of further research for major QTLs related for drought tolerance.
The presence of DNA marker NAU-2954, linked with qtlRWC-1 and qtlELWL on chromosome 23 shows high drought tolerance in cotton. The variety CRIS-134 carries QTLs for drought tolerance and may be selected for breeding programs. These QTLs showed high tolerance against drought stress in cotton.
Acknowledgment
The research work was fully funded by Higher Education Commission Pakistan under Start up Research Grant Project, entitled, “Molecular Tagging of Cotton Genotypes for Drought Tolerance using DNA markers”. The study was also part of thesis work of MPhil student.
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Machado S, Paulsen GM. Combined effects of drought and high temperature on water relations of wheat and sorghum. Plant and Soil, (2001); 233(2): 179-187.
- Tezara W, Mitchell V, Driscoll S, Lawlor D. Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature, (1999); 401(6756): 914-917.
- Lawlor DW, Tezara W. Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: a critical evaluation of mechanisms and integration of processes. Annals of botany, (2009); 103(4): 561-579.
- Burke JJ, O’Mahony PJ. Protective role in acquired thermotolerance of developmentally regulated heat shock proteins in cotton seeds. Journal of Cotton Science, (2001); 5174-183.
- Puspito AN, Rao AQ, Hafeez MN, Iqbal MS, Bajwa KS, et al. Transformation and Evaluation of Cry1Ac+ Cry2A and GTGene in Gossypium hirsutum L. Frontiers in plant science, (2015); 6943.
- Bajwa KS, Shahid AA, Rao AQ, Kiani MS, Ashraf MA, et al. Expression of Calotropis procera expansin gene CpEXPA3 enhances cotton fibre strength. Australian Journal of Crop Science, (2013); 7(2): 206.
- Muzaffar A, Kiani S, Khan MAU, Rao AQ, Ali A, et al. Chloroplast localization of Cry1Ac and Cry2A protein-an alternative way of insect control in cotton. Biological research, (2015); 48(1): 14.
- Yaqoob A, Shahid AA, Samiullah TR, Rao AQ, Khan MAU, et al. Risk assessment of Bt crops on the non‐target plant‐associated insects and soil organisms. Journal of the Science of Food and Agriculture, (2016); 96(8): 2613-2619.
- Yasmeen A, Kiani S, Butt A, Rao AQ, Akram F, et al. Amplicon-based RNA interference targeting V2 gene of cotton leaf curl Kokhran Virus-Burewala strain can provide resistance in transgenic cotton plants. Molecular biotechnology, (2016); 58(12): 807-820.
- Gadallah M. Effect of water stress, abscisic acid and proline on cotton plants. Journal of arid environments, (1995); 30(3): 315-325.
- Wang C-Y, Isoda A, Li M-S, Wang D-L. Growth and eco-physiological performance of cotton under water stress conditions. Agricultural Sciences in China, (2007); 6(8): 949-955.
- Khan MAU, Shahid AA, Rao AQ, Kiani S, Ashraf MA, et al. Role of epicuticular waxes in the susceptibility of cotton leaf curl virus (CLCuV). African Journal of Biotechnology, (2011); 10(77): 17868-17874.
- Schonfeld MA, Johnson RC, Carver BF, Mornhinweg DW. Water relations in winter wheat as drought resistance indicators. Crop Science, (1988); 28(3): 526-531.
- Turner N, Hearn A, Begg J, Constable G. Cotton (Gossypium hirsutum L.): Physiological and morphological responses to water deficits and their relationship to yield. Field Crops Research, (1986); 14153-170.
- Hearn A. The principles of cotton water relations and their application in management; 1994. pp. 14-17.
- Oosterhuis DM. Proceedings of the 2000 Cotton Research Meeting and summaries of cotton research in progress. Special Report-Arkansas Agricultural Experiment Station, (2000); (198).
- Pettigrew W. Physiological consequences of moisture deficit stress in cotton. Crop Science, (2004); 44(4): 1265-1272.
- Yagmur B, Gurel A, Oren Y, Izcl B, Edreva A, et al. Effects of different drought applications and potassium doses on cotton yield and fiber quality. Research Journal of Agricultural and Environmental Management, (2014); 360-67.
- Bartels D. Desiccation tolerance studied in the resurrection plant Craterostigma plantagineum. Integrative and Comparative Biology, (2005); 45(5): 696-701.
- Xoconostle-Cázares B, Ramirez-Ortega FA, Flores-Elenes L, Ruiz-Medrano R. Drought tolerance in crop plants. American Journal of Plant Physiology, (2010); 5(5): 1-16.
- Carter Jr J, Patterson R. Use of relative water content as a selection tool for drought tolerance in soybean; 1985.
- Tahara M, Carver BF, Johnson RC, Smith EL. Relationship between relative water content during reproductive development and winter wheat grain yield. Euphytica, (1990); 49(3): 255-262.
- Bajji M, Kinet J-M, Lutts S. The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regulation, (2002); 36(1): 61-70.
- Paterson A, Saranga Y, Menz M, Jiang C-X, Wright R. QTL analysis of genotype× environment interactions affecting cotton fiber quality. TAG Theoretical and Applied Genetics, (2003); 106(3): 384-396.
- Zhang J, Ma J, Chen X, Liu D, Zhang Z. QTL mapping of fiber quality traits with a composite cross population in upland cotton (Gossypium hirsutum L.). Journal of Agricultural Biotechnology, (2011); 19(2): 230-235.
- Saranga Y, JIANG CX, Wright R, Yakir D, Paterson A. Genetic dissection of cotton physiological responses to arid conditions and their inter‐relationships with productivity. Plant, Cell & Environment, (2004); 27(3): 263-277.
- QIN H-d, ZHANG T-z. QTL mapping of leaf chlorophyll content and photosynthetic rates in cotton. Cotton Sci, (2008); 20394-398.
- SONG XL, GUO WZ, HAN ZG, ZHANG TZ. Quantitative trait loci mapping of leaf morphological traits and chlorophyll content in cultivated tetraploid cotton. Journal of Integrative Plant Biology, (2005); 47(11): 1382-1390.
- Song XL, Zhang TZ. Molecular mapping of quantitative trait loci controlling chlorophyll content at different developmental stages in tetraploid cotton. Plant breeding, (2010); 129(5): 533-540.
- Fan S-l, Yu S-x, Song M-z, Yuan R-h. Construction of molecular linkage map and QTL mapping for earliness in short-season cotton. Cotton Science, (2006); 18135-139.
- Guo Y, McCarty JC, Jenkins JN, Saha S. QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica, (2008); 163(1): 113-122.
- Zhang X, Wang K, Song G, Liu F, Li S, et al. Primary QTL mapping of upland cotton RIL CRI-G6 by SSR marker. Cotton Sci, (2008); 20192-197.
- Babar M, Saranga Y, Iqbal Z, Arif M, Zafar Y, et al. Identification of QTLs and impact of selection from various environments (dry vs irrigated) on the genetic relationships among the selected cotton lines from f 6 population using a phylogenetic approach. African Journal of Biotechnology, (2009); 8(19).
- Liu D, Zhang J, Zhang K, Wang W, Zhang Z. QTL mapping of seed physical traits in upland cotton (Gossypium hirsutum L.). Acta Agronomica Sinica, (2010); 36(1): 53-60.
- Shi L, Hu L, Hu B, Chen L, Wang P. QTL mapping of yield and agronomic traits of interspecific hybrid cotton. Xinjiang Agricultural Sciences, (2010); 47(1): 67-72.
- Zhang P, Zhu X, Guo W, Zhang T. Genetic analysis and QTLs tagging of lint percentage and its closely related yield components in upland cotton. Jiangsu Journal of Agricultural Sciences, (2005); 21(4): 264-271.
- Amjid MW, Malik TA, Shakeel A, Wahid A. QTL Mapping for Relative Leaf Water Contents, Cell Membrane Stability and Excised Leaf Water Loss under Drought by using EST-SSR Markers in Gossypium hirsutum. International Journal of Agriculture & Biology, (2015); 17(4).
- Saleem M, Malik T, Shakeel A, Ashraf M. QTL mapping for some important drought tolerant traits in upland cotton. The Journal of Animal & Plant Science, (2015); 25502-509.
- Saeed M, Guo W, Ullah I, Tabbasam N, Zafar Y, et al. QTL mapping for physiology, yield and plant architecture traits in cotton (Gossypium hirsutum L.) grown under well-watered versus water-stress conditions. Electronic Journal of Biotechnology, (2011); 14(3): 3-3.
- Hanson P, Lu S-F, Wang J-F, Chen W, Kenyon L, et al. Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Scientia Horticulturae, (2016); 201346-354.
- Golestan Hashemi F, Rafii M, Razi Ismail M, Mohamed M, Rahim H, et al. Opportunities of marker‐assisted selection for rice fragrance through marker–trait association analysis of microsatellites and gene‐based markers. Plant Biology, (2015); 17(5): 953-961.
- Mallick N, Sharma J, Tomar R, Sivasamy M, Prabhu K. Marker‐assisted backcross breeding to combine multiple rust resistance in wheat. Plant breeding, (2015); 134(2): 172-177.
- Clarke JM, Townley-Smith TF. Heritability and relationship to yield of excised-leaf water retention in durum wheat. Crop Science, (1986); 26(2): 289-292.
- Clarke JM, McCaig TN. Excised-leaf water retention capability as an indicator of drought resistance of Triticum genotypes. Canadian Journal of Plant Science, (1982); 62(3): 571-578.
- Blum A, Ebercon A. Cell membrane stability as a measure of drought and heat tolerance in wheat. Crop Science, (1981); 21(1): 43-47.
- Doyle JJ. Isolation of plant DNA from fresh tissue. Focus, (1990); 1213-15.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0






