Full Length Research Article
Microscopic detection of chloroplast transgenic plastids using fluorescent probe
Shahid Nazir1,*, Muhammad Sarwar Khan2
Adv. life sci., vol. 5, no. 3, pp. 121-129, May 2018
*- Corresponding Author: Shahid Nazir (Email: shahidnazir97@yahoo.com)
Authors' Affiliations
2- Center of Agricultural Biochemistry & Biotechnology (CABB), University of Agriculture, Faisalabad, Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Fluorescent marker genes have modernized many areas of molecular biology, specifically in plant biotechnology and genetic engineering studies. The use of fluorescent proteins permits the scientists to purify the desired clones visually in transformation work. Green fluorescent protein (gfp) derived from Aequorea victoria has been the most common and favorite fluorescent marker which is being widely used as a visual selection marker gene. It can be easily visualized under UV light without the involvement of any substrate and is non-destructive as well.
Method: A species-specific chloroplast transformation vector was constructed with gfp as a fluorescent marker gene. The recombinant vector was biolistically integrated in tobacco plastome and transgenic cells were initially screened on spectinomycin containing regeneration medium.
Results: The successful plastome integration was verified by using cellular DNA from drug resistant clones in PCR and southern blotting. The expression of gfp in transplastomic clones was microscopically investigated using simple florescent as well as confocal laser scanning microscopes.
Conclusion: Regeneration of transgenic plants was significantly helped by visual identification of fluorescent at different stages of development, also enabling to identify the homozygous and heterozygous tissues. No toxic effect of the gfp was observed and lack of toxicity as maintained by normal phenotypic performance of plants.
Keywords: Fluorescent marker, Microscopy, Chloroplast transformation, Tobacco
Plant genetic transformation is a technique not only for improving the different agronomic characters, but also for basic research of plant genetics, physiology, cellular and molecular biology and pathology etc. [1]. The basic theme of the genetic transformation work is the development of plants with new and improved characteristics. The successful introduction of desirable traits normally involve the development of reliable and competent transformation procedures. All the genetic transformation methods demand separate protocols for integration of targeted gene into living cells, for screening of the transformed cells and regeneration into full plant from transformed cells. Selectable marker gene(s) have been crucial in the development of plant transformation systems because these genes permit researcher to monitor and purify the transgenic from non-transgenic cells [2]. These selectable marker genes are normally co-transformed with targeted gene. There are three types of selection markers (antibiotic, herbicidal and visual), antibiotic resistant and visual reporters are currently used in genetic transformation experiments. For antibiotic selection system, different selectable marker genes like nptII (kanamycin), hpt (hygromycin), Bar is an herbicide PPT tolerant marker (phosphinothricin), aadA (spectinomycin & streptomycin), neo (neomycin) etc. are routinely used in plant transformation work. The respective antibiotics are used in regeneration medium and transformed cells having selectable marker gene can survive and grow, while untransformed cells going to bleach and die due to selection pressure of antibiotic. The utilization of fluorescent probe/visual reporter gene has indication of a new era in the in-vivo examination of sub-cellular events. The availability of an appropriate visible reporter gene is very crucial for improving the plant biotechnology and genetic transformation protocols. A good reporter gene for plant genetic engineering and transformation would be one that is non-destructive and visually score-able without the involvement of any substrate. Furthermore, the reporter gene product should not be harmful to targeted plant tissues and normal plant growth and development [3]. Routinely firefly luciferase (luc), bacterial β-glucuronidase (gus) and jellyfish green fluorescent protein (gfp) etc. are very popular reporter genes and are being used in plant genetic engineering experiments. Among these luciferase and β-glucuronidase both requires the addition of substrate at optimized level to evaluate the enzyme function. In addition the β-glucuronidase assay is destructive and some plant tissues could express an endogenous β-glucuronidase like activity that could be interfere with the identification of transgene product [4]. On the other hand the green fluorescent protein from Aequorea victoria (Jelly fish) produces green fluorescence when illuminated with long-wave ultra violet light [5] and has advantages over other visual markers because fluorescence protein can be visually detected in living cells without the involvement of any substrate and tissue destruction systems. Transformed cells expressing gfp gene, can be isolated very soon after transformation [6]. For optimizing plant transformation protocol, early and strong expression of reporter gene can be helpful and gfp has turned into a commonly used reporter gene [7].
Genetic transformation of chloroplast offers several unique advantages over routinely used nuclear transformation experiments. These includes the absence of transgene escape due to maternal inheritance in most of the agronomic crops, high expression level of foreign proteins due to polyploidy nature, introduction of multiple genes in a single transformation event, absence of gene silencing and position effects due to site-specific integration and homologous recombination etc. [8,9]. Stable genetic transformation requires that all the genome copies should be uniformly transformed to obtain for homoplasmic state. This process requires a gradual screening of transformed plastids during selection. The most competent and commonly used antibiotic marker for chloroplast transformation is spectinomycin [10]. In this study, the species-specific chloroplast transformation vector was constructed with antibiotic resistant (aadA) and reporter (gfp) genes, and was successfully integrated into tobacco plastome via biolistic transformation technique. Further, the transformed cells screened on antibiotic media were microscopically investigated for the presence of fluorescent protein.
Construction of Species-Specific Chloroplast Transformation Vector
The chloroplast transformation vector was developed by amplifying the flanking sequences from the tobacco genomic DNA isolated by CTAB method with modifications [11]. Flanks were PCR amplified using specific primers as described by [12]. Amplified fragment of approximately 2.2kb was cloned into TA cloning vector (Thermo Scientific, Lithuania). The cloned plasmid was restricted with EcoRV (site was engineered with both primers) and ligated in pBlueScriptII restricted with PvuII enzyme. In the resultant clone MCS part of pBlueScriptII was replaced with tobacco flanks. Another SmaI/HincII fragment of MCS carrying ClaI, HindIII, EcoRV, EcoRI and PstI sites was introduced at PvuII site present in-between the tobacco flanks. The final tobacco chloroplast transformation vector was developed by cloning the 2.0kb fragment containing marker gene(s) construct.
Plant Material and Growth Conditions
Sterilized seeds of Nicotiana tabacum var Petite Havana were aseptically germinated on MS solidified medium plates having 2.6 g/L phytagel, 30 g/L sucrose and 4.33 g/L MS salts [13]. The pH of the media was adjusted at 5.8 with the help of 0.1 N KOH and 0.1 N HCl and autoclaved at 121oC for 25 minutes at 15 psi. The plates were placed in incubation room at 25±2oC under 16/8 hours day/night cycle. Three weeks old germinated seedlings were transferred into jars having the same medium for roots and shoots proliferation. Fully matured and healthy green leaves of 6 weeks old tobacco plants were used for chloroplast genetic transformation experiments.
Chloroplast Transformation and Screening of Transgenic Clones
Biolistic genetic transformation system was used to develop chloroplast transgenic plants. The plasmid DNA of final transformation vector was coated on 1.0 µm gold particles as described by [14, 9]. Six weeks old healthy and green tobacco leaves were used in the transformation experiments. After transformation the leaves were chopped into 0.5 x 0.5 mm small pieces and were placed on antibiotic containing regeneration medium for the screening of transformed cells. After six weeks the regenerated shoots on screening medium were shifted for second round of selection and regeneration. The developed shoots were transferred onto rooting medium with same dose of spectinomycin @ 500 mg/L. Genomic DNA was isolated and exposed to PCR for the confirmation of transgene(s) integration [11].
Microscopic Detection of Transformation Events
Transgenic plastids were first analyzed by using Olympus SZX stereomicroscope (Olympus SZX9, Japan) equipped with CCD camera and GFP detection system. Tissue sections of 0.5 x 0.5 cm were placed on a glass slide, 1-2 drops of water was added and then covered with a transparent cover slip. The images developed by the microscope were viewed on attached computer screen. Sub-cellular localization of fluorescent protein was verified by confocal laser scanning biological microscopy (FV1000 Olympus, Japan). Two different types of filters FITC and TRITC were used for gfp and chlorophyll fluorescence respectively. The excitation wavelength for gfp fluorescence was adjusted at 488-514 nm while for chlorophyll it was 560-580 nm. The images produced by gfp and chlorophyll fluorescence were analyzed on computer screen attached with the microscope [14].
Southern Hybridization
Site-specific transgene integration was verified by southern blotting technique. 10 µg of cellular DNA from chloroplast transgenic and untransformed tobacco plants was restricted with ApaI enzyme for overnight at 37oC and separated on 1.0% agarose gel at 60 voltage for 3-4 hours [9]. Gel was washed with depurination, denaturation and neutralization solutions and DNA was transferred on nylon membrane (AppliChem GmbH, Germany) using iBlot Gel Transfer System (Invitrogen, Israel). The BamHI+BglII probe fragment from tobacco chloroplast flanks was hybridized with DNA membrane and detected using Biotin Chromogenic Detection Kit (Thermo Scientific, Lithuania).
Development of Tobacco Chloroplast Specific Transformation Vector having Fluorescent Probe
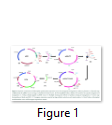
Plant transformation vector is the prerequisite for the development of transgenic plants. The tobacco chloroplast transformation vector was developed by sequential amplification and cloning of inverted repeat region from plastome using primers 5´-GAT ATC AAA ACC CGT CCT CAG TTC GGA TTG C-3´ and 5´-GAT ATC CAC GAG TTG GAG ATA AGC GGA-3´ as mentioned by [12]. These inverted repeats serve as flanking sequences to make easy homologous recombination for carrying exogenous DNA into the tobacco chloroplast genome. Amplified tobacco flanks (TF) was first cloned in the cloning vector (pTZ57R/T) and confirmed with enzymes restriction and colony PCR. pBlueScript II (pBSII) vector was used as the back bone for the development of final chloroplast transformation vector (Fig. 1-A). The multiple cloning sites (MCS) from pBSII were removed with PvuII enzyme and EcoRV restricted fragment of 2.2 kb of tobacco flanks (from pTZ57R/T) was ligated to generate pBSII/TF (Fig. 1-B, C). EcoRV sites were engineered on both ends of the forward and reverse primers used for the amplification of tobacco flanks. TF were containing a PvuII site in-between the trnI and trnA gene sequences and this site was used to clone the MCS to simplify the further cloning of transgene(s). The MCS was ligated with HincII+SmaI blunt ended restriction enzymes at PvuII site to develop pBSII/TF/MCS (Fig. 1-D, E). The expression cassette of aadA and gfp genes was picked from pMSK49 [14] and was introduced into MCS of pBSII/TF/MCS to develop final species-specific chloroplast transformation vector (NTM) having antibiotic and visual selection marker genes (Fig. 1-F, G).
Plastid Transformation using Micro-Projectile Bombardments
Sterile tobacco plants are required to be developed for the genetic transformation of constructs having desirable genes. Fully expanded tobacco leaves being the ideal plant parts with efficient transformation ability were used. For this purpose seeds of Nicotiana tabacum L. var. Petit Havana were surface sterilized and cultured on MS medium plates. Viable seeds germinated and seedlings were propagated in-vitro on phytagel solidified MS-medium in magenta boxes. The vigorous, lush green and completely expanded six weeks old healthy leaves from these in-vitro grown plants were used in transformation experiments. The species specific tobacco chloroplast transformation vector (NTM) was multiplied in E. coli and 1.0 µg of isolated plasmid DNA was coated on 1.0 µm gold micro-carriers. Chloroplast transformation was attempted using the biolistic delivery method following the protocols explained by [9].
In-Vitro Screening and Regeneration of Transformed Tobacco Leaves
Green leaves of Nicotiana tabacum were bombarded with gold micro-carriers carrying the marker genes construct. The leaves were kept on MS medium plates on the Whatman No. 1 filter sheets. After 48 hours of transformation, leaves were chopped into 5×5 mm sections and shifted on regeneration medium supplemented with spectinomycin dihydrochloride @ 500 mg/L. Antibiotic resistant cells started proliferate and shoots were developed, whereas sensitive cells began to bleach as shown in Fig. 2-A. This was the first round of selection of transformed tissues on antibiotic containing medium. The regenerated leaves were again chopped and placed on same selection media for second round of selection and regeneration (Fig. 2-B). The antibiotic resistant shoots recovered were moved onto selective and maintenance medium for root development and propagation of antibiotic resistant shoots (Fig. 2-C). The untransformed plants growing on maintenance media without any antibiotic, demonstrated similar phenotypic performance in comparison with transformed shoots on antibiotic supplemented maintenance media. Hence, no pleiotropic effects of transgene(s) cassette were observed in tobacco plants. The in-vitro developed six weeks old drug resistant plants were exposed to natural conditions for acclimatization and hardening (Fig. 2-D).
Verification of Transgene Integration by PCR
Primary screening of antibiotic resistant clones was carried out using DNA based PCR methodology. Total genomic DNA was isolated from both antibiotic resistant transformed and non-transformed tobacco plants. This isolated and purified DNA was used in PCR for genetic screening of drug resistant shoots. Chloroplast transformation vector (NTM) was mainly composed of antibiotic resistant (aadA), visual marker (gfp) genes and tobacco flanking sequences. Physical map of transformed marker genes cassette with primer position has been shown in Fig. 3-A. Specific primer pairs for aadA (A19/A20) were used to corroborate the presence of antibiotic resistant gene in transformed tobacco plants. Amplification of 552bp fragment confirmed the presence of selection cassette in the antibiotic resistant plants, since no fragment was amplified from non-transformed tobacco (Fig. 3-B). Furthermore, another primer set (GFF/GFR) was used to amplify the reporter gene from aadA positive plants. In Fig. 3-C successful amplification of 743bp fragments of reporter gene also revealing the integration of selection cassette in tobacco as there were no amplification in non-transformed tobacco plants. Further to reconfirm the presence of transgenes cassette, another set of primers pair (A19/GFR) where forward primer lands on aadA gene and the reverse primer from gfp gene was used. Amplification of 1.4kb fragment verified the incorporation of foreign genes into tobacco plant (Fig. 3-D). The absence of the amplified product from the non-transformed plant DNA has further confirmed the transgenic events. However, these experiments only confirm the presence of transgenes in tobacco plant. But to confirm the integration of transgenes into plastome, further experiments were carried out.
Microscopic Detection of Chloroplast Transgenic Plastids
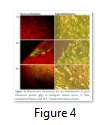
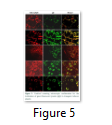
Visible reporter genes are very useful to evaluate the efficiency of newly developed protocols in plant transformation technology. The basic idea is to distinguish transformed regenerants or shoots from heterogeneous population. Biolistic delivery of transformation vector NTM containing gfp as a visual selection marker gene was genetically introduced into tobacco chloroplast. Transplastomic segments in the chimeric leaf tissues can be recognized visually, thus considerably reducing the time and attempts necessary to attain genetically stable plants. The gfp expression yielded fluorescent shoots at an early age making this reporter gene helpful for early identification of transgenic cells. Multiplying transformed cells and the regenerating shoots on selection medium were frequently checked with a hand held long wave UV light for the fluorescence of gfp. Transformation was confirmed by a stereomicroscope equipped for gfp detection system. The size of the fluorescent sectors was different in different leaves in heterozygous transgenic plants depending upon the segregation of the transformed cells from wild type non-transformed tissues (Fig. 4). Under normal light the leaf section will appear green in color. Sub-cellular localization of fluorescent protein was also confirmed by laser-scanning confocal microscopy. This system was comprised on argon mixed gas laser with lines at 488 and 568 nm and detector channels. The channels could be adjusted for green fluorescence and rhodamine images. Green fluorescent protein was detected in FITC detector at 488-514 nm of wavelength, while chlorophyll fluorescence was detected in the TRITC channel using wavelength range 560–580 nm. The images developed by gfp and chlorophyll fluorescence using confocal laser scanning microscopy are given in the Fig. 5. These results also verify the presence of fluorescent protein in the transplastomic tobacco cells. Thus regeneration of transgenic plants was significantly helped by visual recognition of fluorescent at different stages of development, also enabling to identify the homozygous and heterozygous tissues. No toxic effect of the gfp was observed and lack of toxicity was maintained by normal phenotypic performance of plants.
Southern Hybridization for Site-Specific Integration and Homoplasmy
Integration of marker gene(s) cassette in transgenic tobacco chloroplast was confirmed through southern blotting, using flanking sequences as a probe. A 0.8 kb DNA fragment containing tobacco chloroplast flanks, obtained using BamHI+BglII restriction enzymes was used as a probe (Fig. 6-A). Upon restriction of genomic DNA with ApaI restriction enzyme, marker genes transformed tobacco plants were expected to produce one fragment of about 6.0 kb, carrying the transgene(s) cassette (Fig. 6-B) whereas non-transformed chloroplasts expected to show only one band of ~4.0 kb (Fig. 6-C). Restricted genomic DNA was transferred onto nylon membrane and hybridized with 0.8 kb chloroplast specific flanking probe. Fig. 6-D showing the successful hybridization of probe with its target sequences on the membrane. These results indicated the successful incorporation of transgenic cassettes into tobacco plants. Furthermore, it also verified that the marker genes expression cassette has been integrated into the chloroplast genome of tobacco plants and has been attained its homoplasmic state, as no signal appeared against the non-transformed tobacco on the membrane (Fig. 6-D).
Figures
Green Fluorescent Protein (GFP) from Aequorea victoria is a well known protein and has been optimized for higher expression and visual capabilities. Gfp has grown in fame and has been successfully used as a powerful visual marker/reporter in molecular biology and transgenic plant work for more than 20 years. This fluorescent protein has the distinguished capability to transduce blue light to green light; hence, we can visualize gfp fluorescence in genetically engineered plants simply by the exposure of a bright UV light on leaves [15]. In plant transformation studies with the problem of low efficiency, the non-destructive examination of transformed tissues using gfp as reporter gene could significantly help to standardize the transformation procedures for the development of transgenic plants [16].
Tracking of highly expressed genetically transformed events/cells as early as possible is very much attractive for the purpose of keeping them segregated A variety of biotechnologies and nanotechnologies have been developed that could be used to monitor the presence and expression of foreign genes in real-time and in field studies. Nanotechnologies based detection methods comprised on quantum dots, aptamers, molecular bacons etc. need more basic and practical study to reveal that they can possibly be used in living plants in field-based examinations for transgene expression [17]. The biotechnologies used for the identification of transgene or its product continue to move forward. Some primary screening methods like PCR, ELISA and blotting can be easily adopted for existing plant applications. However, these protocols sometime involve sampling and expensive laboratory equipment. Visual identification of transformed tissues is gaining much popularity in genetic transformation experiments as it is very fast and required less expertise as compared to other screening protocols. The purification of transgenic cells with the help of microscopes not only saves the time but also to some extent less expensive. The plant molecular biology community is now using gfp as a routine investigative tool in genetic engineering work.
In present research work, the species-specific chloroplast transformation vector NTM (Fig. 1) was constructed bearing the gfp gene in addition to spectinomycin resistant aadA gene. The vector was biolistically introduced into the tobacco plastome and initial screening of transgenic plastids was carried out on antibiotic containing regeneration medium. The transgene integration was also verified at genomic level using PCR and southern blotting techniques. Higher plant plastid normally contain up to 10,000 plastome copies depends upon the age and tissue type [8] and in the result of genetic transformation very few genome copies get transformed initially. Highly expressed fluorescent probe/marker in plastids facilitates the purification of transplastomic sectors visually from chimeric tissues at early stage of transformation [14]. Drug resistant PCR positive tobacco plants were microscopically investigated to track the fluorescent probe in leaf tissues. Initial screening was done by stereomicroscopy equipped with gfp detection system and chloroplast transgenic plastids were further verified using confocal laser scanning microscope. Gfp is a very powerful and most favorite fluorescent probe used in genetic engineering experiments. It is independent of any destructive methods and does not require any cofactor or substrate for fluorescence. A variety of other bioluminescent probes requires the involvement of other substances before they glow. For example, aequorin require calcium and coelenterazine before fluoresce, and luciferase needs ATP, magnesium and luciferin to glow. This would make gfp a much more versatile tracer probe than others [18]. Moreover, inducible or constitutive expression of gfp can be identified in intact plant tissues [6, 19]. Non-toxic effects on plant cells make it an ideal fluorescent probe for genetic engineering studies.
The authors declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgement
This research work was funded by the Higher Education Commission (HEC) Islamabad, Pakistan. Authors also thanks to Ministry of Science and Technology (MoST) and Pakistan Science Foundation (PSF) for awarding projects to MSK.
- Pawar BD, Jadhav AS, Kale AA, Chimote VP. Strategies for Use of Environment Friendly Alternative Markers for Detection of Transformation Events in Transgenic Plants. Asian Journal of Experimental Biological Sciences, Spl (2010); 1-5.
- Miki B, Mchugh S. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. Journal of Biotechnology, (2004); 107(3): 193-232.
- Kaeppler HF, Menon GK, Skadsen RW, Nuutila AM, Carlos AR. Transgenic oat plants via visual selection of cell expressing green fluorescent protein. Plant Cell Reports, (2002); 19(7): 661-666.
- Hansch R, Koprek T, Mendel RR, Shulze J. An improved protocol for eliminating endogenous β-glucuronidase background in barley. Plant Science, (1995): 105(1): 63–69.
- Haseloff J, Amos B. GFP in plants. Trends in Genetics, (1995); 11(8): 328–9.
- Harper BK, Mabon SA, Leffel SM, Halfhill MD, Richards HA, Moyer KA, Stewart CN. Green fluorescent protein as a marker for expression of a second gene in transgenic plants. Nature Biotechnology, (1999); 17(11): 1125-1129.
- Jordan MC. Green fluorescent protein as a visual marker for wheat transformation. Plant Cell Reports, (2000); 19(11): 1069-1075.
- Daniell H, Khan MS, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends in Plant Science, (2002); 7(2): 84–91.
- Nazir S, Khan MS. Chloroplast-encoded chlB gene from Pinus thunbergii promotes root and early chlorophyll pigment development in Nicotiana tabaccum. Molecular Biology Reports, (2012); 39(12): 10637-10646.
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proceeding of the National Academy of Sciences United Stated of America, (1993); 90(3): 913–917.
- Nazir S, Khan MS. Integration of novel chlorophyll genes from black pine into the chloroplast genome of tobacco. Pakistan Journal of Botany, (2013); 45(S1): 595-600.
- Khan MS, Hameed W, Nozoe M, Shiina T. Disruption of the psbA gene by the copy correction mechanism reveals that the expression of plastid-encoded genes is regulated by photosynthesis activity. Journal of Plant Research, (2007); 120(3): 421–430.
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum, (1962); 15(3): 473–497.
- Khan MS, Maliga P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nature Biotechnology, (1999); 17(9): 910–915.
- Stewart CN. The utility of green fluorescent protein in transgenic plants. Plant Cell Reports, (2001); 20(5): 376–382.
- El-Shemy H, Khalafalla A, Fujita MMK, Ishimoto M. Improvement of protein quality in transgenic soybean plants. Biologia Plantarum, (2007); 51(2): 277–284.
- Stewart CN. Monitoring the presence and expression of transgenes in living plants. Trends in Plant Science, (2005); 10(8): 390-396.
- El-Shemy H, Mutasim A, Khalafalla M, Ishimoto M. The Role of Green Fluorescent Protein (GFP) in Transgenic Plants to Reduce Gene Silencing Phenomena. Current Issues in Molecular Biology, (2008); 11(1): 21–28.
- Richards HA, Han CT, Hopkins RG, Failla ML, Ward WW, Stewart CN. Safety assessment of recombinant green fluorescent protein orally administered to weaned rats. The Journal of Nutrition, (2003); 133(6); 1909-1912.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0