Full Length Research Article
Molecular typing of Methicillin Resistance Staphylococcus aureus (MRSA) isolated from device related infections by SCCmec and PCR-RFLP of coagulase gene
Muhamad Sohail*, Zakia Latif
Adv. life sci., vol. 6, no. 1, pp. 34-40, November 2018
*- Corresponding Author: Muhamad Sohail (Email: drsohailmmg@gmail.com)
Authors' Affiliations
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: SCCmec and PCR-RFLP are productive and cost-effective methods for epidemiological investigations and source tracking of MRSA. Aim of this study was to investigate the epidemiology and molecular characterization of MRSA recovered from device related infection.
Methods: A total of 626 MRSA were collected from prosthetic device related infections and subjected for presence of mec gene and SCCmec typing. SCCmec characterized MRSA were subjected for agr typing followed by RFLP genotyping and amplification of PVL gene. Following the Clinical & Laboratory Standards Institute (CLSI), antibiotic resistance pattern was determined.
Results: Of 626 MRSA isolates, 488(78%) were characterized by SCCmec typing. Most common type was SCCmec IV (43%), followed by SCCmec II (25%) SCCmec III (22%) and only 10% was SCCmec V. SCCmec characterized MRSA strains were 100% resistance to tobramycin and chloramphenicol, 96% resistance to ciprofloxacin and 93% resistance to azithromycin. The gene PVL was only present in SCCmec IV. All agr typed MRSA strains were resistance to gentamicin, tobramycin and chloramphenicol. PVL was present in all strains of agr III, 80% of agrIV, 50% of agrII and absent in agrI. RFLP analysis resulted in 16 types of non-duplicate unique bands pattern which were equally distributed among prosthetic device related infection.
Conclusion: PVL harboring SCCmec or agr typed MRSA strains are less resistance to antibiotics. RFLP is simple, productive and cost-effective method for molecular typing of MRSA. Correct use of antibiotics and molecular surveillance is indispensable to detect the change in epidemiological and antibiotic resistance trends of MRSA.
Keywords: Methicillin Resistant Staphylococcus Aureus (MRSA), Device related infection, SCCmec typing, agr typing, RFLP, Panton–Valentine leukocidin gene (PVL)
Most of device related infections are associated with Staphylococcus aureus, which harbors a lot of toxins enzymes and virulent factors [1]. Device related infections pose major problem toward the patient safety especially when causative agent is methicillin resistant Staphylococcus aureus (MRSA) which is notorious for antibiotic resistance. Standard guidelines of identification and antimicrobial resistance determination were used for better identification and antibiogram of MRSA [2]. Minimum inhibitory concentration (MIC) of vancomycin was assayed using E-test following interpretation criteria of CLSI. Expression of virulent factors of MRSA is under the control of four major types of accessory gene regulator (agr) system [3]. In addition to this agr system, density dependent expression of proteins called quorum sensing is also involved in the pathogenesis and different stages of biofilm formation. The timely and effectively activation of agr system is essential to initiate and establish the infection. This system is also known to produce certain factors to escape the host immune response [4]. Staphylococcal cassette chromosome mec (SCCmec) is a mobile genetic element (21-67 kb) that confers resistance to methicillin, is integrated into chromosome of MRSA. Acquisition of mecA gene causes the alteration of penicillin binding protein (PBP2) gene and makes methicillin, synthetic penicillin, cephalosporins and carbapenems ineffective against MRSA. SCCmec typing is essential to determine epidemiology and evolution. SCCmec genetic element is divided into eight types and subtypes [5]. Efficient typing of MRSA is essential to limit and monitor the spread of pathogen among hospitals and communities. There are many efficient, complex, expensive and time-consuming methods for typing but RFLP getting popularity due to cost effectiveness and simplicity [6]. Coagulation is produced by Staphylococcus aureus essentially and it is a pathogenic and identification. Coagulase gene (coa) was subjected for Restriction Fragment Length Polymorphism (RFLP). MRSA secretes some toxins like PVL which destroys the immune cells and help to tolerate the immune response [7]. Emergence of community acquired MRSA (CA-MRSA) poses major problem to community, on other hand hospital acquired MRSA (HA-MRSA) was threat to hospital admitted, immunocompromised patients.
Aim of this study was to investigate the molecular typing of MRSA isolated from device related infections on the basis of SCCmec typing, agr typing, PCR-RFLP analysis and antibiotic resistance profiles. Selected strains of MRSA were subjected for 16S rRNA amplification followed by gene sequencing.
A total of 626 strains of MRSA were isolated from 6,424 prosthetic devices related infections following standard microbiological guidelines as previously described [8]. These samples were collected from six types of prosthetic devices and catheters including CVC (Central Venous catheters), ETT (Endotracheal Tubes), VA/VP shunts, urinary catheters, orthopedic implants and peritoneal catheters.
Antibiogram: Antimicrobial sensitivity was done by Kirby-Bauer disk diffusion method following the CLSI guidelines [2]. MIC of vancomycin (BioMerieux-USA) was determined by E-test following interpretive criteria of CLSI. Staphylococcus aureus control strain ATCC25923 was used as quality control for antimicrobial sensitivity testing.
DNA extraction: Genomic DNA was extracted from all bacterial cells following the protocol of CTAB/NaCl extraction method with small modifications [9]. Briefly, Overnight culture was centrifuged at 22,000 RCF for 10 minutes to get the pellet. The pellet was suspended in TE buffer and incubated at 37C° for one hour after addition of 10% SDS and proteinase K. After incubation NaCl and CTAB/NaCl was added, mixed well and incubated at 65C° for 10 minutes. Upper most layer was collected and mixed with chilled phenol/chloroform/isoamyl alcohol and centrifuged at 1400rpm for 10 minutes. Supernatant was collected and mixed with isopropanol to get DNA precipitated. After centrifugation DNA pallet was washed with 70% alcohol and dissolved in 50ul of deionized water.
Molecular detection of mecA and PVL gene: Amplification of mecA and PVL gene was done in all MRSA strains. MRSA were confirmed by the presence of mecA gene. Amplification of mecA was performed with primers mecA-F (5'GTGAAGATATACCAAGTGATT 3') and mecA-R (5'ATGCGCTATAGATTGAAAGGAT3') [10]. Presence of PVL gene is detected with primers Luk-PV-F (5'ATCATTAGGTAAAATGTCTGGACATGATCCA3') and Luk-PV-R (5'GCATCAAGTGTATTGGATAGCAAAAGC3') [11].
SCCmec typing: Multiplex PCR was used to amplify the six types of SCCmec gene using the set of primers already reported [12]. Multiplex PCR for subtypes of SCCmec IV was performed separately and all others in the same reaction. The reaction mixture of 50μl contained master mixture, template DNA and primers.
agr grouping: agr grouping was performed by multiplex PCR using primers reported in 2015 [12]. This PCR grouped MRSA strains in four categories. There were some strains which are not classified by these primers and denoted as agr0.
Restriction Fragment Length Polymorphism (RFLP) analysis: Bacterial genome DNA was extracted and coa gene was amplified using the previously described set of primer and conditions of amplification [13]. Amplified product was digested using two restriction enzymes (aluI and HaeII; NEB England). Restriction was done following the manufacturer guidelines in the same reaction tube. After restriction digestion gel electrophoresis was done with 3% agarose to detect the restriction fragments.
16S rRNA sequencing: Using the universal primers 785F (GGATTAGATACCCTGGTA) and 907R (CCGTCAATTCMTTTRAGTTT) 16S rRNA was amplified and sequenced from Macrogen, Korea. The sequences obtained were BLAST in National Centre for Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast) for identification of bacterial strains and observed similarity to reported sequences. The sequences obtained from Macrogen were submitted to GeneBank for accession numbers. After successful submission accession numbers were obtained.
Phylogenetic tree: After getting accession number from GeneBank, a phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7) software on basis of similar sequences in NCBI database keeping bootstrap value 1000 [14].
A total of 626 isolates were confirmed as MRSA by amplification of mecA gene, isolated from prosthetic device related infections. Of 626 isolates of MRSA, 98% were resistance to ofloxacin, 97% were resistance to ciprofloxacin, 92% were resistance to tobramycin, 88% were resistance to erythromycin, 88% were resistance to azithromycin and erythromycin and 87% resistance to gentamycin. None of isolate was resistance to vancomycin and linezolid [15].
SCCmec characterization: The distribution of SCCmec determined by multiplex PCR. Out of 626 strains, 22% (138/626) didn’t belong to any group. SCCmec IV was most common group (43%) among MRSA followed by type II 25%, type III 22%, type V 10%. A total of 488 strains were typed by SCCmec typing method (Figure 1). Prevalence of antibiotic resistance among various SCCmec characterized MRSA is given in the Table 1. Results of antimicrobial resistance showed variable resistance pattern among different groups of SCCmec typing. Overall, all SCCmec groups were resistant to tobramycin and chloramphenicol; sensitive to vancomycin and linezolid, other antibiotics showed variable resistance among different groups.
agr typing
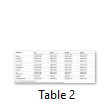
Distribution of agr genes determined among MRSA isolated form prosthetic devices. Some of MRSA, 14% (68/488) didn’t belong to any agr group and denoted as agr0 group. The most frequent type was agr type I which was 61% followed by agr type IV 16%, agr type II 13% and agr type III 10% (Figure 1). Out of 488 isolates of MRSA 420 were types by agr typing method, remaining 68 isolates didn’t belong to any group. Distribution of antimicrobial resistance among different agr types is given in the Table 2. All of 55 isolates belonged to agr II, were resistance to all antibiotics except vancomycin and linezolid. Similarly, 67 isolates belonged to agr IV were resistance to all tested antibiotics except vancomycin and linezolid. Other two groups (agr I and agr III) showed the variable resistance towards tested antibiotics.
Restriction Fragment Length Polymorphism: A total of 420 strains of MRSA were typed by SCCmec and agr typing, were subjected for PCR-RFLP analysis. coa gene was amplified by specific primers. Amplified products were digested with HaeII and AluI restriction enzymes. On the basis of pattern of bands from both restriction enzymes 16 unique types of band pattern were identified (Figure 2). One representative for each unique pattern was selected for further investigations. Antibiogram, biofilm potential, SCCmec typing, agr typing was similar for all pathogens showing same banding pattern.
Panton-Valentine leukocidin (PVL) gene characterization: The PVL gene was detected in SCCme and agr typed MRSA. The PVL genes was found only in SCCmec IV typed MRSA. All other SCCmec don’t have PVL gene. On other hand PVL gene was present in 100% agr type III, 80% of agr type IV and 50% of agr type II.
Phylogenetic analysis: On the basis of antimicrobial sensitivity pattern, SCCmec typing, agr typing 420 strains of MRSA were selected. Which were subjected for PFLP analysis and resulted in 16 unique non-duplicate DNA band patterns. Sixteen MRSA isolates were selected form each unique DNA pattern for 16S rRNA sequencing. Among selected strains, nine were HA-MRSA and seven were CA-MRSA.Phylogenetic tree was constructed separately for CA-MRSA and HA-MRSA because these two groups had similarity among them. Phylogenetic tree showed selected strain of CA-MRSA (ZS28C, ZS35C, ZS38C, ZS41C, ZS43, ZS59C and ZS64C) were 99% to 100% similar to the reported sequences. Similarly, HA-MRSA (ZS39H, ZS40H, ZS42H, ZS44H, ZS45H, ZS46H, ZS47H, ZS48H and ZS49H) were 99 to 100 % similar to the reported sequences. Phylogenetic tree of CA-MRSA and HA-MRSA with accession number is given in the Figure 3 and 4 respectively.
Tables & Figures
Most of catheter related infections are due to MRSA which causes community acquired and hospital acquired infections. Source identification is very important while dealing with outbreak of infectious disease. Most authentic and valuable tool for source identification is molecular typing [16]. Pulse Field Gel Electrophoresis (PFGE) and Multi Locus sequence typing (MLST) are standard methods for tying of S. aureus but these are expensive and laborious [17].
This study was designed to do the molecular typing of MRSA isolated from prosthetic device related infections and determine their antibiotic profile. None of MRSA was characterized as SCCmec I, the most prevalent type was SCCmec IV (43%) followed by SCCmec II (25%), SCCmec III (22%) and SCCmec V (10%). These results were consistent with previous studies describing SCCmec IV most prevalent followed by type III, V and I [18]. Some of MRSA (24%) isolated form prosthetic devices were not typed by agr system. Among typed MRSA 61% belonged to agr type I followed by agr IV, agr II and agr III, these results were similar to the findings reported in Iran in 2017[18]. The difference in the prevalence of agr and SCCmec typing with some studies was due to source of specimens [19-22]. PCR-RFLP was done to genetically type the MRSA isolated from prosthetic device related infections and sixteen strains were selected for further testing. Different studies showed different DNA banding patterns with same set of enzymes. The variations were due to sample source and geological location, but the similarity was that all DNA bands were multiple of 80bp [23,24].
All SCCmec types isolates of MRSA were resistance to tobramycin and chloramphenicol and sensitive to linezolid and vancomycin which correlates the antimicrobial sensitivity testing conducted in Iran [25,26]. For aminoglycosides (amikacin, gentamicin and tobramycin), azithromycin some studies showed similar results [27] and other showed different results [28]. SCCmec IV harboring PVL was less resistance to the antibiotics compared to the SCCmec I, SCCmec II, SCCmec III and SCCmec V that don’t have PVL gene. MRSA belonged to agr III were less resistant to antibiotics as compared to agr I that don’t have agr typing gene. These results showed that PVL gene was not associated with antibiotic resistance especially in the MRSA isolated form prosthetic devices. MRSA isolates harboring PVL were less resistant to the antibiotics [29].
All SCCmec IV have PVL gene and similar results of association of SCCmec IV and PVL reported in China [30]. MRSA types as SCCmec IV was the major MRSA that spreads from community to the hospital setting and same results reported in Libia [27]. The study described that PVL gene is present in 40% of MRSA isolated from prosthetic device related infection, these results were similar to the study conducted in Oman (54%) and different from the results reported in Kuwait hospital (14.6%) [31,32]. MRSA containing PVL gene were most commonly associated with skin and soft tissue infection which justified the high prevalence of PVL containing MRSA from prosthetic devices.
In Conclusion, MRSA isolated form device related infections were more resistant to antibiotics than any other type of infection. SCCmec IV along with PVL and agr I was more prevalent genotype of MRSA involving device related infections.
The authors declare that there is no conflict of interest regarding the publication of this paper.
- Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. New England Journal of Medicine, (2004); 351(16): 1645-1654.
- CLSI (2015) M100-S26. Performance Standards for Antimicrobial Susceptibility Testing: 26th Informational supplement. Clinical and Laboratory Standards Institute Wayne, PA.
- Peerayeh SN, Azimian A, Nejad QB, Kashi M. Prevalence of agr specificity groups among Staphylococcus aureus isolates from university hospitals in Tehran. Laboratory Medicine, (2015); 40(1): 27-29.
- Wright JS, Jin R, Novick RP. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proceedings of the National Academy of Sciences of the United States of America, (2005); 102(5): 1691-1696.
- Wong H, Louie L, Lo RY, Simor AE. Characterization of Staphylococcus aureus isolates with a partial or complete absence of staphylococcal cassette chromosome elements. Journal of clinical microbiology, (2010); 48(10): 3525-3531.
- Arakere G, Nadig S, Swedberg G, Macaden R, Amarnath SK, et al. Genotyping of methicillin-resistant Staphylococcus aureus strains from two hospitals in Bangalore, South India. Journal of clinical microbiology, (2005); 43(7): 3198-3202.
- DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infectious Disease Clinics, (2009); 23(1): 17-34.
- Sohail M, Latif Z. Prevalence and antibiogram of methicillin resistant Staphylococcus aureus isolated from medical device-related infections; a retrospective study in Lahore, Pakistan. Revista da Sociedade Brasileira de Medicina Tropical, (2017); 50(5): 680-684.
- Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol, (2001); Chapter 2Unit 2.4.
- Makgotlho PE, Kock MM, Hoosen A, Lekalakala R, Omar S, et al. Molecular identification and genotyping of MRSA isolates. FEMS Immunology & Medical Microbiology, (2009); 57(2): 104-115.
- McClure J-A, Conly JM, Lau V, Elsayed S, Louie T, et al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant staphylococci. Journal of clinical microbiology, (2006); 44(3): 1141-1144.
- Nelson MU, Bizzarro MJ, Baltimore RS, Dembry LM, Gallagher PG. Clinical and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit in the decade following implementation of an active detection and isolation program. Journal of clinical microbiology, (2015); 53(8): 2492-2501.
- Himabindu M, Muthamilselvan DS, Bishi DK, Verma RS. Molecular analysis of coagulase gene polymorphism in clinical isolates of methicillin resistant Staphylococcus aureus by restriction fragment length polymorphism based genotyping. American Journal of Infectious Diseases, (2009); 5170-176.
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, (1985); 39(4): 783-791.
- Sohail M, Latif Z. Prevalence and antibiogram of methicillin resistant Staphylococcus aureus isolated from medical device-related infections; a retrospective study in Lahore, Pakistan. Revista da Sociedade Brasileira de Medicina Tropical, (2017); 50: 680-684.
- Havaei SA, Ghanbari F, Rastegari AA, Azimian A, Khademi F, et al. Molecular typing of hospital-acquired Staphylococcus aureus isolated from Isfahan, Iran. International scholarly research notices, (2014); 185272.
- Weller T. Methicillin-resistant Staphylococcus aureus typing methods: which should be the international standard. Journal of Hospital Infection, (2000); 44(3): 160-172.
- Nasirian S, Saadatmand S, Goudarzi M. Molecular typing of methicillin-resistant Staphylococcus aureus strains isolated from patients admitted to intensive care units of hospitals in Tehran city, Iran, based on agr pattern. Journal of Isfahan Medical School, (2017); 35(451): 1424-1431.
- Bayat B, Zade MH, Mansouri S, Kalantar E, Kabir K, et al. High frequency of methicillin-resistant Staphylococcus aureus (MRSA) with SCCmec type III and spa type t030 in Karaj's teaching hospitals, Iran. Acta Microbiologica et Immunologica Hungarica, (2017); 64(3): 331-341.
- Perovic O, Singh-Moodley A, Govender NP, Kularatne R, Whitelaw A, et al. A small proportion of community-associated methicillin-resistant Staphylococcus aureus bacteraemia, compared to healthcare-associated cases, in two South African provinces. European Journal of Clinical Microbiology and Infectious Diseases, (2017); 36(12): 2519-2532.
- Udo EE, Boswihi SS. Antibiotic Resistance Trends in Methicillin-Resistant Staphylococcus aureus Isolated in Kuwait Hospitals: 2011-2015. Medical Principles and Practice, (2017); 26(5): 485-490.
- Vali L, Dashti AA, Mathew F, Udo EE. Characterization of heterogeneous MRSA and MSSA with Reduced Susceptibility to chlorhexidine in Kuwaiti hospitals. Frontiers in Microbiology, (2017); 8(JUL).
- Eed EM, Ghonaim MM, Hussein YM, Saber TM, Khalifa AS. Phenotypic and molecular characterization of HA-MRSA in Taif hospitals, Saudi Arabia. The Journal of Infection in Developing Countries, (2015); 9(03): 298-303.
- Nada T, Ichiyama S, Osada Y, Ohta M, Shimokata K, et al. Comparison of DNA fingerprinting by PFGE and PCR-RFLP of the coagulase gene to distinguish MRSA isolates. Journal of Hospital Infection, (1996); 32(4): 305-317.
- Goudarzi H, Seyedjavadi SS, Udo EE, Beiranvand E, Fazeli M, et al. Molecular Characterization and Distribution of Class 1 Integron-Bearing Methicillin Resistant Staphylococcus aureus Strains in Burn Patients, Tehran, Iran. Jundishapur Journal of Microbiology, (2017); 10(2): e40592.
- Lu Z, Chen Y, Chen W, Liu H, Song Q, et al. Characteristics of qacA/B-positive Staphylococcus aureus isolated from patients and a hospital environment in China. Journal of Antimicrobial Chemotherapy, (2014); 70(3): 653-657.
- Khemiri M, Alhusain AA, Abbassi MS, El Ghaieb H, Costa SS, et al. Clonal spread of methicillin-resistant Staphylococcus aureus-t6065-CC5-SCCmecV-agrII in a Libyan hospital. Journal of global antimicrobial resistance, (2017); 10: 101-105.
- Mohammadi S, Sekawi Z, Monjezi A, Maleki M-H, Soroush S, et al. Emergence of SCCmec type III with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare-and community-acquired infections in the west of Iran. International Journal of Infectious Diseases, (2014); 25: 152-158.
- Bhatta DR, Cavaco LM, Nath G, Kumar K, Gaur A, et al. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC infectious diseases, (2016); 16(1): 199.
- Qin Y, Wen F, Zheng Y, Zhao R, Hu Q, et al. Antimicrobial resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates from child patients of high-risk wards in Shenzhen, China. Japanese journal of infectious diseases, (2017); 70(5): 479-484.
- Monecke S, Skakni L, Hasan R, Ruppelt A, Ghazal SS, et al. Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC microbiology, (2012); 12(1): 146.
- Udo EE, Al-Lawati B-H, Al-Muharmi Z, Thukral S. Genotyping of methicillin-resistant Staphylococcus aureus in the Sultan Qaboos University Hospital, Oman reveals the dominance of Panton–Valentine leucocidin-negative ST6-IV/t304 clone. New microbes and new infections, (2014); 2(4): 100-105.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0