Full Length Research Article
Trackable CEMB-Klean Cotton Transgenic Technology: Affordable Climate Neutral Agri-biotech Industrialization for Developing Countries
Zahida Qamar1,*, Muhammad Tariq1, Tahir Rehman1, Muhammad Shahzad Iqbal1, Muhammad Bilal Sarwar1, Muhammad Nauman Sharif1, Zohaib Hassan1, Ayesha Ahmad2, Aiman Zahra3, Ayesha Latif4, Bushra Rashid1, Mohsin Abbas Zaidi5, Bushra Tabassum1, Sameera Hassan1, Allah Baksh6, Momina Javaid1, Sania Akram1, Saira Azam1, Farah Naz1, Shafique Ahmed7, Kamran Shahzad Bajwa8, Mudassar Fareed Awan7, Naila Shahid1, Arfan Ali9, Saman Riaz1, Bisma Bashir1, Kanza Sadiq1, Qurat ul Ain Kokab1, Iqra Yousaf1, Abdul Munim Farooq1, Muhammad Aslam Javed10, Zia Ur Rahman11, Muhammad Zafar Saleem1, Aneela Yasmin1, Muhammad Umar Bhatti12, Usman Arif13, Khurram Bashir14, Arshad Jamal15, Shahid Javed Butt9, Asif Arif16, Irshad Ahmad17, Abdul Qayyum Rao1, Muhammad Saleem Haider18, Tassawar Hussain Malik19, Idrees Ahmad Nasir1
Adv. life sci., vol. 6, no. 3, pp. 131-138, May 2019
*- Corresponding Author: Zahida Qamar (Email: florilab5@gmail.com)
Authors' Affiliations
2. Plant protection, Yuman Agricultural University, Kuming 65021, Yunnan, China.
3. Virtual University of Pakistan, Multan, Pakistan.
4. Plant protection, Yuman Agricultural University, Kuming 65021, Yunnan, China.
5. University of Ottawa, 451 Smyth Road, Ottawa ON K1H 8M5, Canada.
6. Agricultural Genetic Engineering, Faculty of Agricultural Sciences and Technologies, Nigde Omer Halisdemir University, 51240, Nigde-Turkey.
7. Department of Molecular Biology and Biochemistry, University of Management and Technology, Sub-Campus, Sialkot, Pakistan.
8. Chinese Academy of Agricultural Sciences, Beijing China.
9. FB Genetics (Pvt) Ltd, Four Brothers Group Pakistan, Lahore, Pakistan.
10. Agribiotechnology Research Institute, Ayub Agriculture Research Institute, Faisalabad, Pakistan.
11. Cardiff University UK.
12. University of Arizona USA.
13. Department of Biotechnology, University of the Central Punjab, Lahore, Pakistan.
14. Plant Genomic Network Research Team, CSRS, RIKEN, Yokohama, Tsurumi-Ku, Kanagawa, 230-0045, Japan.
15. Department of Biology, College of Science, University of Ha’il, Kingdom of Saudi Arabia.
16. Molecular Cell Biology of Plants Ludwig-Maximilians-University of Munich, LMU, Germany.
17. Department of Life Sciences, King Fahd University of Petrolium and Minerals, Dhahran 31261, Saudi Arabia.
18. Institute of Agriculture Sciences, University of the Punjab, Lahore, Pakistan.
19. Central Cotton Research Institute, Multan · Pakistan Central Cotton Committee (PCCC), Old Shujaabad Road, Multan, Pakistan.
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Transgenic technology reflects the incorporation of novel useful traits in crop plants like cotton for economic benefits by overcoming the problems including insects’ pests and weeds in special. The present study is the success story of the continuous effort of CEMB team started back in the 1990s.
Methods: This study includes characterization of a large number of Bacillus thuringiensis (Bt) strains taken from local soil and subjected to direct transformation of isolated BT genes into local cotton cultivars. Protocols for transformation into cotton plants were optimized and validated by the development of double gene codon optimized (Cry1Ac and Cry2A) transgenic cotton varieties.
Results: The resulting GMOs in the form of CEMB-33, CA-12, CEMB-66 have been approved by Punjab Seed Council in 2013 and 2016 respectively. Double Bt and weedicide resistant cotton harboring CEMB-Modified and codon optimized cp4EPSPS (GTGene). These varieties can tolerate glyphosate spray @ 1900ml per acre without the appearance of necrotic spots/shedding and complete removal of all surrounding weeds in the cotton field is a significant advance to boost cotton production without spending much on insecticides and herbicides.
Conclusion: In the current report, two unique sets of primers which amplify 1.1 Kb for CEMB-double Bt genes and 660 bp product for CEMB-Modified cp4EPSPS (GTGene) were tested. CEMB cotton variety CKC-01 is specially designed as low cost and easy to use by local farmer’s technology has the potential to revolutionize the cotton growing culture of the country.
Keywords: Event detection; Bt Cotton; CEMB transgenic technology; GTGene
Cotton crop is the main fiber source and an important cash crop of Pakistan that provides raw material to the textile industry [1]. It contributes up to 60% of the foreign exchange annually, with 8.2% of the value added in agriculture that counts about 2% to total GDP [2]. Pakistan is the 4th top producer of cotton in the world, yet in terms of yield, it is at the distant 10th position [3,4]. Biotic stress factors like an attack of insect pest, an infestation of the cotton leaf curl virus diseases and weeds are the keys factors responsible for the lower cotton production [5,6]. In Pakistan, approximately more than 150 species of chewing and sucking insect pests attack different stages of cotton and caused loses up to 2.5 million bales annually [3]. Cotton bollworm (Helicoverpa armigera), pink bollworm (Pectinophora gossypiella), spotted bollworm (Earias insulana), and armyworm (Spodoptera littoralis) fall in chewing category while whitefly (Bemisia tabaci), thrips (Thrips tabaci), jassid (Amrasca devastans), and aphids (Aphis gossypii) are mainly included in sucking category [7]. Traditionally the insect pest management mainly relies on the use of chemicals of worth $8.1 billion per annum globally and out of these, nearly $2.7 billion are spent only for cotton pest management. Non-judicious and frequent use of pesticide not only results in the development of the resistance in the targeted insect pest but also indirectly effects human health as well as environment.
Weeds infestation is another challenge for cotton crop production that accounts for 37% of yield loss annually [2,8,9]. Weeds not only compete with the cultivated plants for water, sunlight, and available nutrients but also provides shelter and food for plant pathogens. Mechanical and chemical methods including manual hoeing and herbicide spray are time-consuming, labor-intensive and expensive ways of weeds removing. Up until now, many conventional strategies have been devised to incorporate the blood from existing germplasm that all end up with little success due to the lack of resistant gene pool in available genotypes. The use of advanced technologies provides us an opportunity to genetically engineered the plant with enhanced protection, production and with greater nutritional value in a possibly short time [2,7,10,11]. These genetic modifications break the sexual incompatibilities barrier among the cross-species and enormously increase the size of the available gene pool from the prokaryotic or viral origin [12].
Bacillus thuringiensis is a Gram-positive spore-forming bacterium with entomopathogenic properties and a long history of safe use as a sprayable biopesticide [13]. Parasporally formed crystals are predominantly composed of one or more proteins, which penetrate epithelial cells of the insect midgut by inserting pores into the plasma membrane. These proteins also are known as Bt toxin which is harmful to the larvae of moths, butterflies, beetles, and flies. When insects feed on the plant, the toxin enters the body and binds to the insect’s gut. Hence, it disrupts its feeding and digestion process and eventually leads to the death of the insect. Various studies and field trials on the crop plants that harbor the Cry portions in their leaves from Bacillus thuringiensis have shown the effectiveness as an alternative to the synthetic insecticides [9,10,14]. Glyphosate (N-phosphono methyl glycine), is a non-selective potent herbicide which interferes in the shikimate metabolic pathway by inhibiting the synthesis of 5-enolpyruvyl- 3-phosphoshikimate (EPSPS). This disruption prevents the synthesis of the three important aromatic amino acids, phenylalanine, tryptophan, and tyrosine and is subsequently leads to plant death (Herrmann and Weaver, 1999). Stable transformation of the bacterial gene encoding EPSPS protein in the plant can make the plant resistant against glyphosate weedicide spray. Currently, the most widely accepted genetically modified traits in GM crops are herbicide tolerance (HT) and insect resistance (IR). GM soybean, maize, canola, and cotton are the most common examples of these crops in the market [15,16]. Currently, USA is on the top of the list of world GMO crop producer with 70.9 million ha of cropland followed by Brazil (44.2 million hectors), Argentina 24.3 million ha and India 11.6 million and Canada 11.0 million ha respectively. The use of GM cotton plant in India has registered phenomenal growth in cotton production and topped the world with 95% resilient adoption rate [7].
Keeping in view the potential of above-mentioned genetic resources, for the first time in Pakistan a transformation of Cry1Ac and Cry2A in cotton was started in early 2000. Although the efficiency and characterization of these Cry proteins had studied been studied and confirmed by the CEMB researchers in the early 90s [17-20]. A single construct harboring the CEMB Cry1Ac+Cry2A driven by CaMV35S promoter, and nptII gene was transformed in local cultivar CIM-482 of Gossypium hirsutum. Later, Rashid et al., reported the stable advanced event of this transformation and name as CEMB-02, which got NBC approval in 2009 and subsequent commercialization license in 2016. Azam et al., [21] reported the dissemination of CEMB-02 derived Bt cultivar on all-over the cotton growing belt of Pakistan that raised the local farmer’s profit mostly by low or no seed cost decreasing the pesticides expenditures to control the pest attack [22]. CEMB has also been continuously conducting risk assessment studies since 2000 of the CEMB 02 event harboring the double gene Bt on different organisms including Mice, Fish, Earthworm, Chicks and Rabbits along with gene flow studies including horizontal and vertical gene flow which results in delaying their efforts to be brought in front of public [21].
CEMB started work on the development of herbicide tolerance in Gossypium hirsutum to save the yield losses by weed infestation. [8] reported the stable transformation of synthetic codon-optimized 5-enolpyruvylshikimate-3-phosphate synthase gene cloned into pCAMBIA 1301 vector under a 35S promoter with Agrobacterium tumefaciens in CEMB-02 event having the Cry1Ac+Cry2A genes in a single cassette. They also further evaluated the transgenic line harboring the Cp4 EPSP transgene line by glyphosate spray at the concentration of 1900ml/acre. one of the advance lines that pass the spray and insect bioassay with stable transgene expression was named as CEMB-Klean Cotton. In this study, to address the regulatory and effective labeling issues, we reported the PCR based detection of the CEMB-Klean cotton event by using the two set of unique and specific primers pairs and also confirmed the through the ELISA for protein quantification of the transgene.
Testing Material and Tissue Sampling
The 40 days old grown homozygous lines of CEMB-Klean cotton transgenic plants as testing material were obtained from CEMB cotton research substation Multan for molecular analysis. leaf samples; newly emerged leave from the top; mature leaf from the middle; and older leaf from the bottom of six plants were cleaned with ddH2O, harvested with a sterile scissor from each plant separately and immersed into liquid nitrogen. The leaves of non-transgenic cultivar CIM-482 were used as control samples. These samples were then ground well into a very fine powder and stored at -80°C till further molecular investigations.
Isolation of gDNA and PCR Analysis of CEMB-Klean Cotton Plants
In 2019 it does not make much sense to write protocols in such a detail naming the protocol would be enough. In my opinion, the DNA isolation and PCR analysis should be combined and shortened significantly.
Modified CTAB based DNA isolation method was used with slight modification as mentioned by the Shah et al. Briefly as, the grounded samples were mixed with pre-heated CTAB buffer having freshly added 1% in ratio Mercaptoethanol, incubated at 65°C for thirty min with vigorous shaking on vortex mixer each after 5 min. A pre-chilled equal volume of Chloroform-Isoamyl alcohol (24:1) (sigma#C0549-1QT) was added to the incubated samples and mixed thoroughly by inverting the tubes. For supernatant separation centrifugation of the mixed samples was carried out at 10K rpm for 30 min at 4°C. The obtained supernatant was mixed with double volume of pre-chilled ethanol (Merk # 64-17-5) and 1/10 volume of 3M sodium acetate (pH:5.2) (sigma# S7899-100ML), incubated at -70°C for 5 h for genomic DNA precipitation. For pellet formation, the incubated samples were further centrifugated at 12k rpm for 30 min at 4°C. gDNA pellet washing was carried out by 70% ethanol and purified by the genomic DNA purification kit (Cat#: K0512) by Thermo Scientific as per manufacturer protocol. Total RNA contamination from the extracted DNA was removed by using the RNase A solution by the Thermo Scientific (Cat# EN0531) as per manufacturer protocol. The DNA integrity was accessed by the gel electrophoresis while Quantity and purity were accessed by the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc) by measuring the absorbance at 260nm (Table-1).
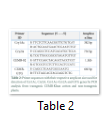
The specific amplification of the CEMB CryAc+Cry2A and GTG genes from the CEMB-Klean cotton plants were carried out by using the specifically designed primers. Table 2 shown the primer sequences with their excepted amplicon size. These primers were designed by using the Primer Premier 6.0 primer designing software and synthesized from the Eurofins Oligo synthesis facility Pvt. Ltd. The total 25µl reaction volume was prepared for a single conventional PCR reaction using the 10XPfu+MgSO4 PCR reaction buffer (Thermo Scientific™ cat# 10202830). Veriti™ 96-well Thermal Cycler (Thermo Scientific Cat#4375786) was programmed as; initial denaturation for 3min at 95°C, followed by the 35 cycles with cyclic condition includes denaturation 95°C for 30 sec, annealing at 52°C for CEMB-02 primer and 51°C for GTG primer for 30 sec, followed by the cyclic extension at 72°C for 30 sec. At the last stage, a final step of the final extension for 10 min at 72°C was added. The amplified products were further resolved on 1.5% agarose gel prepared in the 1x TAE buffer, stained with “ethidium bromide” and visualized by gel documentation system (UVP GelTower: Analytik Jena the US).
ELISA Immunoassay of CEMB-Klean Cotton Plants
Enzyme-linked immunosorbent assay (ELISA) was carried out to determine the expression of CEMB Cry1Ac+Cry2A and GTG genes at the protein level. Young, mature and old leaves tissues from the top, middle, and bottom of CEMB-Klean cotton plants were harvested, crushed well in liquid nitrogen and incubated on ice for 1 hour in 1ml protein extraction buffer [1]. Extracted crude protein was harvested by the centrifugation of the incubated samples at 13k rpm for min at 4°C. Gene-specific protein quantification was carried out by using the Envirologix ELISA kit (Cat. # 051) by following the manufacturer’s protocol.
Genomic DNA Extraction and Quality Evaluation
The Modified (Cetyltrimethylammonium-bromide) CTAB method followed by the column-based purification was used to extract the good quality of DNA from the well ground stored samples. The extracted yield of DNA from all the samples was in the range of 520 to 800 ng/µl. Qualitative 260/280 and 260/230 ratios for all the samples were in the range of 1.85 to 2.01 (Table 1). The integrity and quality of the extracted DNA with intact bands are shown in Figure 1.
PCR Based Detection of CEMB Cry1Ac+Cry2A and GTG Genes with Specific Primers
The CEMB-Klean cotton transgenic plants contain locally isolated Cry1Ac and Cry2A genes in a single construct [14] and GTG gene as a separate event in a different construct that was transformed and evaluated previously [8]. In this study, we targeted the CEMB-Klean Cotton transgenic plants and CIM-482 non-transgenic plants to check the integration of the stable transgene at the DNA level by using specific primers. Initially, an amplification of the Cry1Ac and Cry2A genes was carried out by gene-specific primers. The Cry1Ac specific primer for Cry1Ac Bt gene and Cry2A specific primer for Cry2A Bt gene amplified both genes independently and recorded the 382 bp and 730 bp amplification respectively (Figure 2). The CEMB-02 primer generated the 1.1kb specific product from all tested samples. The CEMB-GTG primer amplified the 660 bp band from the transgenic plants (Figure 3). Both these primers gave the unique amplification for the CEMB Klean-cotton harboring transgene while no amplification was observed from the control plants. Therefore, both these primer (CEMB-02 and CEMB-GTG) could be used to screen out the CEMB-Klean cotton transgenic plants from mixed populations. No non-specific amplification was observed by these primers on the gel (Figure 2-3).
Transgenes Protein Estimation Through ELISA
Enzyme-linked immunosorbent assay (ELISA) procedure was used to detect the protein concentration of the Cry1AC, Cry2A, cp4EPSP transgene in the transgenic plants. The quantification of the proteins encodes by the studied transgenes are shown in Figure 4. The concentration of Cry1Ac ranged from 2-95 to 3.5 ng g-1 total soluble protein, the concentration of cry2A ranged from 2.8 to 3.8 ng g-1 total soluble protein, while the concentration of cp4EPSP ranged from 2.8 to 3.2 ng g-1 total soluble protein in different transgenic lines (Figure 4).
Figures & Tables
The agriculture sector is the main contributor to the GDP of Pakistan. Cotton is one of the main cash crops, provides raw material to the textile and edible oil industry and considered as a backbone of the economy [11]. Farmers bear the major losses in the production of cotton in recent past due to biotic and abiotic factors like an attack of insect pests, weeds infestation and viruses attack [4]. The use of genetic engineering able the researcher to establish the novel environment-friendly agricultural practices to manage the yield affecting factors [7]. The effective way is to develop the genetically engineered crop plants with improved insect resistance and herbicide tolerance characteristics to reduce the yield losses by an insect pest, labor cost and chemical application [1,2,9-11]. Isolation and characterization of the most efficient gene is prerequisite for crop improvement through genetic engineering. Cry proteins are endotoxin in nature encodes by the soil bacterium Bacillus thuringensis and used to provide resistance against the chewing insect pest [24]. [20,25] carried out the intensive characterization of the Cry proteins from locally isolated strains, record their effectiveness against the insect pests and established an efficient protocol to transform the transgenes via Agrobacterium-mediated transformation method in crop plants in the early 1990s at Center of excellence in molecular biology (CEMB), University of the Punjab, Lahore, Pakistan. Based on these studies Cry1Ac and Cry2A genes were locally isolated cloned in a single cassette and transformed in the Gossypium hirsutum plants in the early 2000s. The stable expression of these genes in the transgenic cotton plants has shown the high mortality rate of chewing insect in homozygous lines due to Synergist effects of Cry1Ac+Cry2A [14]. The combined effect of the endotoxin encoding by more than one gene not only provides the effective protection layer against the insects but also diminishes the chance of resistance development towards the toxin among the insect population [33].
Weeds are the unwanted plants that cause 30% losses in cotton plants by consuming the available moisture, light, and nutrients [8]. These also harbor and provide shelter to insect pest on off season [11]. The conventional way of weeds management is expensive and laborious. Glyphosate (N-phosphonomethylglycine) is a broad-spectrum herbicide that inhibits the 5-enolpyruvyls-Shikimate-3-phosphate (EPSP) synthase pathway. This inhibition ceases the production of aromatic amino acids like phenylalanine, tryptophan, and tyrosine and halts plants growth [2]. The integration and overexpression of bacterial origin glyphosate tolerant gene (GTGene) able the plants to resist the glyphosate spray by encoding the 5-enolpyruvylshikimic acid-3-phosphate synthase (EPSPS) enzyme. [8] initially reported the stable transformation of GTGene in the Gossypium hirsutum transgenic cultivar (CEMB-02) at CEMB and evaluated under glyphosate spray effectively. Later on [2,10] also shown the GTGene efficiency for herbicide tolerance in transgenic cotton plants. The efficiently engineered crop plants should contain a single copy of the transgene in each cell [7]. The detection of the transgene(s) and its quantification in the genetically engineered organism is critical for acceptance and commercialization [34]. The proper detection system to trace the GM contents in the crop plants, molecular characterization of transgene, point of insertion and localization information are the mandatory requirements for their release in many countries like members of EU, India, Australia, China, Japan, Korea, Russia, Taiwan and other before commercialization [35]. The strict implementation by the Cartagena protocol and Convention on Biological diversity also demand specific detection system and adequate level of protection layers before the handing and transfer of the GM materials [35-37]. With an increasing number of product types and possible chance of mixing of GMOs, products have created the demand for the development of specific detection methods. To address the requirements of the regulatory bodies and commissions related to GM crops, DNA based detection methods are applied [7]. Detection of the transgene in the genetically modified plant by polymerase chain reaction is considered as a gold standard due to its sensitivity, cost-effectiveness and user-friendly ability [38]. Even a traceable amount of transgene can be detected by this approach. A number of reports cited the effectiveness of PCR approach to detect the transgene in cotton [11], maize, soybean, canola [39], rice [40] and GM sugar beet [41]. Here in this study, we establish a PCR protocol to detect the CEMB-Klean Cotton transgenic plants by using specific primers. The practice of this protocol with testified primers will support to fulfill the regulatory framework and policies before commercialization.
Here in this study we developed a protocol and reported the two unique set of primers, that detect the CEMB-Klean Cotton transgenic plants specifically by using the conventional polymerase chain reaction. This easy screening method could distinguish CEMB-02/CEMB-Klean Cotton and would be useful to monitor the spread of gene in mixed populations. Monitoring the protein concertation would help researchers to tackle silencing issues if any in proceeding generations. The CEMB-Klean Cotton will help cotton growers to get the benefit of locally developed low-cost technology.
Acknowledgement
The authors are thankful for the Higher Education Commission (HEC) of Pakistan for the provision of the funds and necessary facilities for conducting GM Cotton research.
The authors declare that there is no conflict of interest regarding the publication of this paper.
- Shahid AA, Shaukat MS, Bajwa KS, Rao AQ, Husnain T. Detection of Transgenes in Cotton (Gossypium hirsutum L.) by Using Biotechnology/Molecular Biological Techniques. International Journal of Biological, Biomolecular, Agricultural, Food and Biotechnological Engineering, (2015); 9(1): 87-92.
- Puspito AN, Rao AQ, Hafeez MN, Iqbal MS, Bajwa KS, et al. Transformation and Evaluation of Cry1Ac+Cry2A and GTGene in Gossypium hirsutum L. Front Plant Sci, (2015); 6(943): 1-13.
- Abdullah A. An analysis of Bt cotton cultivation in Punjab, Pakistan using the agriculture Decision Support System (ADSS). AgBioForum, (2010); 13(3): 274-287.
- Spielman DJ, Zaidi F, Zambrano P, Khan AA, Ali S, et al. What are farmers really planting? Measuring the presence and effectiveness of Bt cotton in Pakistan. PloS one, (2017); 12(5): e0176592.
- Asi MR, Afzal M, Anwar SA, Bashir MH. Comparative Efficacy Of Insecticides Against Sucking Insect Pests of Cotton. Pakistan J Life Soc Sci, (2008); 6(2): 140-142.
- Farooq A, Farooq J, Mahmood A, Shakeel A, Rehman KA, et al. An overview of cotton leaf curl virus disease (CLCuD) a serious threat to cotton productivity. Australian Journal of Crop Science, (2011); 5(13): 1823-1831.
- Kamle M, Kumar P, Patra JK, Bajpai VK. Current perspectives on genetically modified crops and detection methods. 3 Biotech, (2017); 7(3): 1-15.
- Latif A, Rao AQ, Khan MA, Shahid N, Bajwa KS, et al. Herbicide-resistant cotton (Gossypium hirsutum) plants: an alternative way of manual weed removal. BMC Res Notes, (2015); 8(1): 1-8.
- Naqvi RZ, Asif M, Saeed M, Asad S, Khatoon A, et al. Development of a Triple Gene Cry1Ac-Cry2Ab-EPSPS Construct and Its Expression in Nicotiana benthamiana for Insect Resistance and Herbicide Tolerance in Plants. Front Plant Sci, (2017); 8(55): 1-9.
- Muzaffar A, Kiani S, Khan MA, Rao AQ, Ali A, et al. Chloroplast localization of Cry1Ac and Cry2A protein–an alternative way of insect control in cotton. Biol Res, (2015); 48(1): 14.
- Sarwar MB, Batool F, Rashid B, Aftab B, Hassan S, et al. Integration and expression of heat shock protein gene in segregating population of transgenic cotton for drought tolerance. Pakistan Journal of Agricultural Sciences, (2014); 51(4): 1-11.
- Cressey D. Transgenics: A new breed. Nature, (2013); 497(1): 27-29.
- Dong H, Li W. Variability of endotoxin expression in Bt transgenic cotton. Journal of Agronomy and Crop Science, (2007); 193(1): 21-29.
- Rashid B, Saleem Z, Husnain T, Riazuddin S. Transformation and inheritance of Bt genes in Gossypium hirsutum. Journal of Plant Biology, (2008); 51(4): 248-254.
- Pray CE, Naseem A. Supplying crop biotechnology to the poor: opportunities and constraints. The Journal of Development Studies, (2007); 43(1): 192-217.
- Dill GM. Glyphosate-resistant crops: history, status and future. Pest Manag Sci, (2005); 61(3): 219-224.
- Riazuddin S, Husnain T, Khan E, Khanum F. Insect resistant transgenic basmati rice. Rice Biotechnology Quarterly, (1995); 23(1): 7-8.
- Husnain T. Transformation of indica rice with synthetic cry1Ac gene. Biologica, (1998); 44(1): 180-192.
- Maqbool SB, Husnain T, Riazuddin S, Masson L, Christou P. Effective control of yellow stem borer and rice leaf folder in transgenic rice indica varieties Basmati 370 and M 7 using the novel δ-endotoxin cry2A Bacillus thuringiensis gene. Molecular Breeding, (1998); 4(6): 501-507.
- Husnain T, Asad J, Maqbool SB, Datta SK, Riazuddin S. Variability in expression of insecticidal Cry1Ab gene in Indica Basmati rice. Euphytica, (2002); 128(1): 121-128.
- Azam S, Samiullah TR, Yasmeen A, ud Din S, Iqbal A, et al. Dissemination of Bt cotton in cotton growing belt of Pakistan. Adv life sci, (2013); 1(1): 18-26.
- Nazli H, Sarker R, Meilke KD, Orden D (2010) Economic performance of Bt cotton varieties in Pakistan. Denver, Colorado Agricultural and Applied Economics Association’s 2010 AAEA, CAES & WAEA Joint Annual Meeting.
- Höfte H, Whiteley HJM, Reviews MB. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev, (1989); 53(2): 242-255.
- Islam R, Malik T, Husnain T, Riazuddin S. Strain and cultivar specificity in the Agrobacterium-chickpea interaction. Plant Cell Rep, (1994); 13(10): 561-563.
- Husnain T, Malik T, Riazuddin S, Gordon MPJPc, tissue, culture o. Studies on the expression of marker genes in chickpea. Plant Cell, Tissue and Organ Culture, (1997); 49(1): 7-16.
- Husnain TJB. Transformation of indica rice with synthetic cry1Ac gene. (1998); 44180-192.
- Chaudhry B, Yasmin A, Husnain T, Riazuddin SJPMBR. Mini-scale genomic DNA extraction from cotton. Plant Molecular Biology Reporter (1999); 17(3): 280-280.
- MAJEED A, HUSNAIN T, Riazuddin S. Transformation of virus-resistant genotype of Gossypium hirsutum L. with pesticidal gene. Plant Biotechnology, (2000); 17(2): 105-110.
- Allah B, Rao A, Khan G, Bushra R, Shahid A, et al. Insect resistance studies of transgenic cotton cultivar harboring cry1Ac and cry2A. CAB Direct, (2012); 5(2): 167-171.
- Pandey A, Kamle M, Yadava L, Muthukumar M, Kumar P, et al. Genetically modified food: its uses, future prospects and safety assessments. Biotechnology, (2010); 9(4): 444-458.
- Delaney B. Safety assessment of foods from genetically modified crops in countries with developing economies. Food Chem Toxicol, (2015); 86(2015): 132-143.
- Fraiture M-A, Herman P, Taverniers I, De Loose M, Van Nieuwerburgh F, et al. Validation of a sensitive DNA walking strategy to characterise unauthorised GMOs using model food matrices mimicking common rice products. Food Chemistry, (2015); 173(15): 1259-1265.
- Fraiture MA, Herman P, Lefevre L, Taverniers I, De Loose M, et al. Integrated DNA walking system to characterize a broad spectrum of GMOs in food/feed matrices. BMC Biotechnol, (2015); 15(76): 1-11.
- Zhang X, Tang Q, Wang X, Wang Z. Structure of exogenous gene integration and event-specific detection in the glyphosate-tolerant transgenic cotton line BG2-7. PLoS One, (2016); 11(7): e0158384.
- James D, Schmidt AM, Wall E, Green M, Masri S. Reliable detection and identification of genetically modified maize, soybean, and canola by multiplex PCR analysis. J Agric Food Chem, (2003); 51(20): 5829-5834.
- Mäde D, Degner C, Grohmann LJEFR, Technology. Detection of genetically modified rice: a construct-specific real-time PCR method based on DNA sequences from transgenic Bt rice. European Food Research and Technology, (2006); 224(2): 271-278.
- Mannerlöf M, Tenning P. Screening of transgenic plants by multiplex PCR. J Plant Molecular Biology Reporter, (1997); 15(1): 38-45.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0