Full Length Research Article
Confirmation of root-knot nematode resistant gene Rmi1 using SSR markers
Musarrat Ramzan*, Rifat Z. Ahmad, Naheed Kauser, Anis Ali Shah, Rabia Saba, Iqtidar Hussain, Shahina Fayyaz, Saifullah Khan
Adv. life sci., vol. 4, no. 2, pp. 55-59, February 2017
*- Corresponding Author: Musarrat Ramzan (Email: marain79@gmail.com)
Authors' Affiliation
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: The Root Knot Nematode (RKN) is a serious economic threat to various cultivated crops worldwide. It is a devastating pest of soybean and responsible to cause severe yield loss in Pakistan. The cultivation of resistant soybean varieties against this pest is the sustainable strategy to manage the heavy loss and increase yield. There is an utmost need to identify RKN resistant varieties of soybean against cultivated in Pakistan. The presented study is an attempt to identify and confirm the presence of resistant gene Rmi1 in soybean.
Method: Molecular studies have been done using Simple Sequence Repeat (SSR) marker system to identify resistant soybean varieties against Root Knot Nematode (RKN) using fifteen (15) indigenous cultivars and four (4) US cultivars. DNA was isolated, purified, quantified and then used to employ various SSR markers. The amplified product is observed using gel documentation system after electrophoresis.
Results: Diagnostic SSR markers Satt-358 and Satt-492 have shown the presence of Rmi1 gene in all resistance carrying genotypes. Satt-358 amplified the fragment of 200 bp and Satt-492 generated 232 bp bands in all resistant genotypes. This study confirmed the Rmi gene locus (G248A-1) in all internationally confirmed resistant including six (6) native varieties.
Conclusion: These investigations have identified six (6) resistant cultivars revealing the effective and informative sources that can be utilized in breeding programs for the selection of RKN resistance soybean genotypes in Pakistan.
Key words: Rmi1 gene, Soybean, Root-knot nematodes, SSR markers, Pakistan
Introduction
Southern root-knot nematode Meloidogyne incognita (Mi) is a serious pest of soybean or Glycine max (L.) and responsible to cause severe yield loss of soybean crop worldwide, particularly in the southern USA and Pakistan [1,2]. The most successful method for controlling the yield losses is the use of Mi resistant soybean genotypes. DNA marker assisted screening of genotypes is known to accelerate the identification of root-knot nematode resistant soybean cultivars. A major quantitative loci (QTL) near the top of the linkage group O (LG-O) confers resistance to Mi and can be identified by DNA markers [1]. Several molecular markers have been used to assess genetic diversity and also to locate/confirm the resistant genes against nematodes including SSR markers, refer to tandem repeat sequences of variable length, and exhibit a high degree of allelic distinction [3,4]; result is abundant and highly reproducible [5]. In soybean, various scientists have identified the locus of resistance to Mi nematode and several recent studies have confirmed this resistance locus against Mi in soybean [6,7]. Tamulonis et al., found that resistance was quantitatively inherited with seed character conferring the resistance locus/ primer G248A-1 linked with Mi resistance in soybean by applying several SSR markers [7].
The objective of this study was to determine the presence of Mi resistant gene present on linkage group O (LG-O) of Pakistani soybean cultivars by applying reported microsatellite markers.
Methods
Collection of plants materials
Total nineteen (19) varieties of soybean were used for present study which includes fifteen (15) Pakistani soybean genotypes and four US cultivars. These indigenous fifteen soybean cultivars were previously screened against Meloidogyne incognita nematode (Mi) in green house and characterized on the basis of resistance and susceptibility to Mi in pathogenicity test (unpublished data).
DNA isolation
Total genomic DNA was isolated according to the modified and optimized CTAB method reported by Doyle and Doyle [8].
Estimation of quality and purity of DNA
Quantification and purity of the DNA was estimated as described by Sambrook and Russell (2001). DNA concentration was measured in spectrophotometer using 1:100 dilutions. Readings were noted for each DNA sample at wavelength 260nm and 280nm. DNA concentration in the sample was calculated by the following formula,
DNA concentration (µg/ml) = A 260×50×dilution factor (100)
Simple Sequence Repeats (SSRs) analysis
On the basis of the genetic linkage map of soybean [9], three SSR markers (Satt_358, Satt_132 and Satt_492) located near Mi resistance QTL on LG-O [10] were selected for this study. Primer sequences for each SSR were obtained from SoyBase, a USDA-sponsored genome database (http://soybase.org/ssr.html). All primers were synthesized by Integrated DNA Technologies, Inc, USA.
DNA amplification using SSRs
DNA amplification reactions were carried out using 3 pairs of SSR primers previously developed for soybean (USDA). Polymerase chain reactions were performed in 20μl reaction mixture containing 8ul PCR reaction mix (Promega) ,the PCR reaction mix contains PCR buffer, 25mM MgCl, 200mM each dNTPs, 1.5 units of Taq polymerase, 2μl (05 picomole/μl)of each reverse and forward primers and 3 ul (50ng/μl) genomic DNA. The PCR amplification programme was as follow: initial denaturation 940C for 03 min, denaturation 35 cycles with 940C for 30 Sec, annealing 460C for 01 min, extension 720C for 01 min and final extension was set 720C for 05 min. PCR was performed using 96 well Eppendorff Master gradient cycler. The amplified product stored at 4ºC for Metaphore agarose gel electrophoresis.
Metaphor agarose (Ultra high-resolution agarose)
Metaphor agarose gel was prepared by addition of 1 mg metaphor agarose and 1mg agarose in chilled 1x TBE (pH 8) buffer. This mixture was melted in a microwave until the agarose dissolved completely and the solution became clear. Ethidium bromide 5µl (0.05µg/ml) was mixed properly in the agarose solution. Once the molten gel solidified after being poured into a cast, it was kept at 4ºC for 20 minutes before use to obtain a good resolution. The electrophoresis was performed in large submarine units (Thermo EC-320) at 60V for 2hr (Thermo EC, EC-250-90). Then gels were removed from the electrophoresis tank and placed in gel documentation system (UV TechTM, UK), the gel images were captured. The size of PCR products were determined by DNA ladder (Fermentas). 100bp ladder (Life TechnologiesTM) used as molecular size marker or gene ruler on each gel.
Data analysis
Amplified PCR products of microsatellite were scored qualitatively for presence and absence for each marker allele-genotype combination. Data were entered into a binary matrix as discrete variables, 1 for presence and 0 for absence of the character. Polymorphism information content (PIC) value of a marker was calculated according to formula [11]. Only major bands consistently amplified were scored and faint bands were not considered. All computations were carried out using the NTSYS-PC, version 2.2 packages [12].
Results
For this study 3 SSR markers (Satt_358, Satt_132 and Satt_492) were selected based on the integrated soybean genetic linkage map. These markers are reported to be located near the G248 A-1 diagnostic marker for Mi-resistance. Satt _358 exhibits 200 bp amplified band in Mi resistant homozygous genotype, 200/192 bp bands in heterozygous Mi resistant genotypes and 192 bp band in Mi susceptible genotypes. Satt_132 is reported to amplify characteristic 238 bp band in Mi resistant cultivars whereas in susceptible cultivars it could be 236, 246, 248, 250 or 252 bp band. Satt_492 is reported to amplify 232 bp bands in resistant as well as susceptible soybean cultivars.
In PCR results of US Mi resistant soybean genotypes Forrest and Gregg and US Mi susceptible genotypes Lee and Bossier were used as control and confirmed a major resistant Mi locus gene near diagnostic markers (G248 A-1) with Satt_358 and Satt_132 markers which amplified expected band size (similar to the resistant genotype). This confirmed the presence of G248 A-1 on the linkage group of O in the positive tagged genotypes.
The entire studied soybean cultivars with Satt_358 marker amplified either 200 bp fragment or 192 bp band except one cultivar NARC-1 which did not show any amplified band. 10 cultivars showed 200 bp band and 7 cultivars exhibited 192 bp band. The microsatellite marker Satt_132 amplified 238 bp except in Rawall-1genotype.
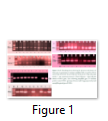
Among susceptible cultivar 250 bp fragment was amplified in six genotypes and absent in PR-142 genotype while these entire bands were also present in reported resistant and susceptible cultivars. For Satt_492 marker, all studied genotypes showed 2 banding patterns except NARC-3, Ajmeri and NARC-2. 7 genotypes showed 232 bp fragments whereas 9 genotypes exhibited 250 bp band fragments (Table 1; Fig. 1).
Tables & Figures
Discussion
In the present study microsatellite marker technique was utilized to screen indigenous soybean germ plasm for RMi resistance. SSR markers Satt_358 and Satt_132 have been reported to identify resistant genotypes whereas Sat_492 has been found to be less effective. Bo et al. and Li et al. used six SSR markers flanking the G248 A-1 locus on LG-O to track the inheritance of this locus in different soybean lines [1,10]. Bo et al., also proved in his study that Mi resistance in soybean cultivars was due to the presence of Rmi1 gene on QTL (G248 A-1) on LG-O. They reported strong evidence of co-segregation of RKI resistance and a 200 bp band at Satt_358 marker.
Results of pathogenicity test (data not shown) were relevant to this molecular analysis. Co-descent analysis of markers and phenotype (pathogenicity) showed that Mi-resistant cultivars possess a 200 bp band at Satt_358 and a 238 bp at Sat_132 against southern root-knot nematode (Meloidogyne incognita). The relationship between Mi reaction and Satt_358 and Satt_132 genotypes was previously described, where Mi resistant cultivars inherited a 200 bp (Satt_358) and 238 bp (Satt_132) band and Mi-susceptible cultivars inherited a 192 bp band at Satt_358 (Table 1) [1]. In the present study all tested Mi-resistant cultivars exhibited the 200 bp band for Satt_358 which is in accordance with previous reports [2,7].
Sat_492 is another closest microsatellite marker to Rmi1 gene on LG-O and located near the RMi locus (G248 A-1 locus) on the linkage group O [9]. Tamulonis et al., reported a major G248 A-1 diagnostic locus which was indicated in the Satt_492 to Satt_358 interval was 3.1cM [2]. Results from Mi reactions differ from Satt_492 in 5 out of 7 susceptible genotypes which showed 250 bp band except Ajmeri and NARC-2 genotypes while this 250 bp band is also present in indigenous resistant genotypes except one genotype soybean 95086 which contain 232 bp band, the positive control genotypes exhibited 232 bp band.
RMi allele present in all resistant genotypes except 95086 and susceptible genotypes. The marker Satt_492 was not as strongly associated with Mi resistance as Satt_358 or Sat_132 (Table 1). During our Mi-screening experiment at molecular level, we noticed that 95086 did not exhibit any resistance related band whereas it shows the resistance against Mi during pathogenicity test at green house. This variation in 95086 cultivar contrasting to other tested genotypes might be due to parentage that both have homozygous alleles for this gene.
Genotype 95086 possesses a 200 bp (Satt_358), 238 bp (Sat_132) and 232 bp at Satt_492. On the basis of these results, we concluded that there are two ancestral sources of Mi resistance in this indigenous cultivar. Therefore, in conclusion the tight linkage of both Satt_358 and Sat_132 to the diagnostic marker G248 A-1 on LG-O in studied soybean cultivars indicates that selection for the Mi-resistant allele by any of these markers should be highly effective in identifying Mi-resistant plants / genotypes. Therefore, indigenous resistant germplasm can be effectively used in developing breeding strategies for the selection of Mi- resistant plants.
It is interesting that all Mi-susceptible cultivars possess a 192 bp band at Satt_358 diagnostic marker and 250 bp band at Sat_132. These results strongly support the association of Satt_358 and Satt_132 markers with the G248 A-1 locus for Mi resistance. Thus, the Mi-reaction of these genotypes was predicted on the basis of the presence or absence of the 200 bp and 238 bp allele for Mi-resistance at Satt_358 and at Satt_132, respectively.
In present study utilizing SSR markers, the efforts were extended to screen the targeted resistance gene against southern root knot nematode (M. incognita) located on LG-O, this result is an approach, focusing on the fixing of resistance gene in high yielding genotypes of indigenous soybean germplasm.
References
- Ha B-K, Bennett JB, Hussey RS, Finnerty SL, Boerma HR. Pedigree analysis of a major QTL conditioning soybean resistance to southern root-knot nematode. Crop Science, (2004); 44(3): 758-763.
- Tamulonis J, Luzzi B, Hussey R, Parrot W, Boerma H. DNA markers associated with resistance to Javanese root-knot nematode in soybean. Crop Science, (1997); 37(3): 783-788.
- Panaud O, Chen X, McCouch S. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Molecular and General Genetics, (1996); 252(5): 597-607.
- Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, et al. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theoretical and Applied Genetics, (2000); 100(5): 697-712.
- Gutierrez M, Patto MV, Huguet T, Cubero J, Moreno M, et al. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theoretical and Applied Genetics, (2005); 110(7): 1210-1217.
- Luzzi B, Boerma H, Hussey R. Resistance to three species of root-knot nematode in soybean. Crop Science, (1987); 27(2): 258-262.
- Tamulonis J, Luzzi B, Hussey R, Parrott W, Boerma H. RFLP mapping of resistance to southern root-knot nematode in soybean. Crop Science, (1997); 37(6): 1903-1909.
- Doyle JJ. Isolation of plant DNA from fresh tissue. Focus, (1990); 1213-15.
- Cregan P, Jarvik T, Bush A, Shoemaker R, Lark KG, et al. An integrated genetic linkage map of the soybean genome. Crop Science, (1999); 39(5): 1464-1490.
- Li Z, Jakkula L, Hussey R, Tamulonis J, Boerma H. SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theoretical and Applied Genetics, (2001); 103(8): 1167-1173.
- Meehan D, Xu Z, Zuniga G, Alcivar-Warren A. High frequency and large number of polymorphic microsatellites in cultured shrimp, Penaeus (Litopenaeus) vannamei [Crustacea: Decapoda]. Marine Biotechnology, (2003); 5(4): 311-330.
- Rohlf F. NTSYS-PC 2.02. Numerical taxonomy and multivariate analysis system Applied Biostatistics, Setauket, New York, (1998).