Full Length Research Article
Sequence Diversity of MAOA Gene within Wild and Docile Animal Species
Rashid Saif1, 2,*, Beenish Tariq3, Naila Naz3
Adv. life sci., vol. 5, no. 3, pp. 135-142, May 2018
*- Corresponding Author: Rashid Saif (Email: rashid.saif37@gmail.com)
Authors' Affiliations
2- Decode Genomics, 264-Q, Johar Town, Lahore, Pakistan
3- Department of Biotechnology, Kinnaird College for Women, Lahore, Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Molecular characterization of MAOA gene was performed to investigate aggressive behaviour within wild (lion, leopard, and wolf) as opposed to docile animal (sheep, goat) species living in different habitats, by undertaking sequence diversity analysis of this gene.
Methods: The MAOA gene was partially amplified by PCR for wild and docile animal species. Amplified DNA was sequenced and then analyzed using BioEdit and Sequencher softwares, while multiple sequence alignment and phylogenetics analysis were conducted through MEGA software. Bioinformatics tool like Prosite scan, Motif Scan and Prot Param were used to study properties of mutant proteins of MAOA gene.
Results: Different polymorphic sites were observed which included c.956, c.1063 in docile animals and c.2530 in wild animals. Phylogenetic analysis based on this candidate gene endorsed the existing taxonomy of subject animals, while bioinformatics tools explored the altered characteristics of mutant MAOA protein.
Conclusion: The newly found polymorphic loci in wild and docile animals in this study could have a role in behavioral response and acclimatization within their peculiar habitats. This study also highlights the genetic diversity of MAOA gene, which will add knowledge to the existing animal genetic resource of Pakistan.
Keywords: Sequence diversity, MAOA gene, Wild animal, Docile animal
MAOA enzyme involved in oxidative deamination reactions, which is responsible for controlling behavioral attributes of the animals by degrading the neurotransmitters [1]. This gene is located on X chromosome with total of fifteen exons in Capra hircus (goat) as well as in Ovis aries (sheep). Its product is also found in liver, placenta and gastrointestinal tract. Serotonin, a hormone controlled by this gene is a neuromodulator that is linked with wide range of physiological functions in the central nervous system such as mood swings (mostly good moods) [2]. Absence of this gene has a negative impact leading to depressive behavioral patterns.
This gene has been selected as it greatly influences the behavior of living being. Presence of this protein regulates serotonin while its absence, down regulation or loss of functioning of this gene through manipulation or mutational event may lead to aggression, anxiety, fear, substance abuse and irregular sexual maturation [3]. Mostly, wild animals have much aggression as compared to domestic animals. This is because of their nature to acclimatize themselves according to their environment but genetic factor also plays role in controlling behavioral moods of subject species. Purpose of current study is to characterize and find out the MAOA gene variants contribution with the aggressive and docile behavior of the animals [4].
We obtained blood and skin samples for DNA extraction through proper channel from wild animals like leopard, markhor, wolf and tiger from the zoo veterinarians during treatment procedure and markhor samples from license holder safari agents. Similarly, docile animals of goat and sheep blood were collected from the local farmers and government livestock farms to search out whether the aggressive behavior of these animals has any relationship with the variants of this gene. We follow this approach by comparing functional domain encoding exons of this gene in various species to identify changes that would help them to adapt to their natural habitat. In-silico characterization was also performed on MAOA protein considering these genetic variants [5].
Sample Collection
Samples of wild animals (Rawal Tiger, Sam Tiger, Mohini Tigress, Wild Leopard and Markhor) were collected from Lahore zoo and license holder agents respectively, while samples of docile animals (Mungli sheep, Lehri, Angora, Kamori and Teddy goat breeds) were collected from local farmers and breeder. 5-10 mL of blood was obtained from jugular vein of each selected animal in EDTA containing Falcon tube [6].
DNA Extraction
DNA was extracted using standard inorganic method [7]. Later on, DNA concentration was also estimated through agarose gel electrophoresis at 80 volts for 40 minutes and standard concentration of 50ng/uL of DNA was used for downstream process.
Primer Designing
Gene ID 101106401 of MAOA has 15 exons having its major domain on exon 8 and 15. While, ENSOART00000003319 identifier from ensemble depicts that flavin amine oxidase domain is also very important in docile animals. Primer3 tool was used to design the primers sets for the partial amplification of aforementioned docile animal accession and XM_007084354.1 for wild animals.
PCR optimization, DNA amplification and sequencing
PCR was run on wild and docile animal extracted DNA by using standard protocol for 35 cycles. Primer made for docile species was optimized at 61Cᵒ whereas primer made for wild species was optimized at 59Cᵒ. After it, PCR products were precipitated using EXOSAP (Exonuclease shrimp alkaline phosphatase) and sequenced using Genetic analyzer 3130xL [8].
Bioinformatics tools
All the sequences were aligned using online tools Blast2Sequence (www.ncbi.nlm.nih.gov), and for studying the mutational events from aligned sequences, Bioedit was used [9,10]. Phylogenetic analysis was performed using MEGA (Molecular Evolutionary Genetics Analysis) software. For studying characteristics of mutated protein in the respective species such as post translational modifications, Prosite scan tool was used which generated consensus pattern of that post translated site. To study physio-chemical properties Prot Param tool was applied whereas conserved domains were observed by using Motif Scan [9,11].
Mutational Analysis
After aligning the sequences from Sequencher software and NCBI Blast2Sequence tool, the resulting trimmed cleaned DNA bases were compared to that of reference genes of domestic animals Ovis aries and capra hircus genomes. The Coding DNA sequence was compared with the exon 8 in reference sample. Mutational analysis identified 6 haplotypes in domestic species. Mutant-1 of MAOA comprises of a deletion at position c.956 and polymorphism at position c.1017. This variant was only present in Mungli sheep sample-01. Mutant-2 of MAOA gene comprises of a deletion at position c.956. It is present in Mungli sheep sample-2 alone. Mutant-3 comprises of polymorphic loci at three different positions i.e. c.956, c.1012 and c.1063. It is only present in Lehri goat. Mutant-4 of MAOA gene comprises of a polymorphism at c.956 positions. It is present in two goat breeds only i.e. Kamori and Angora goat. Mutant-5 composed of polymorphisms at c.956 and c.1013 positions. It is present in 2 goat breeds i.e. Kamori-2 and Teddy-2. Mutant-6 of this gene comprises of polymorphisms at several positions including c.992, c.996, c.1002, c.1004, c.1005, c.1007, c.1011, c.1012 and c.1099. It is present only in Teddy goat-1.
For sequence comparison in wild species, the chosen reference sample was Panthera tigris. Upon aligning sequences from wild species exon 15 of Panthera tigris. Five different haplotypes were found. Mutant-1 of MAOA gene in wild species comprises a polymorphism at c.956 positions. It is present only in wild wolf. Mutant-2 with a polymorphism at position c.1063. It is present only in Markhor. Mutant-3 comprises of a mutation at c.2530 position in two species i.e. Tiger Rawal and Leopard. While mutant-4 comprises a mutation at position c.2528 in tiger Sam only. Mutant- 5 appeared with a mutation at position c.2528 and c.2543 in Mohini Tigress.
Phylogenetic Analysis
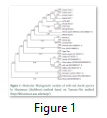
Phylogenetic tree was constructed using Molecular Evolutionary and Genetic Analysis Software (MEGA). This helped in determining the taxonomical order and relation of one species with the other on the basis of partial MAOA gene sequencing by considering main functional domain of this protein. Constructed cladogram gives the insight about mutational events and closely related species on the basis of these polymorphisms. This phylogenetic analysis involved 13 nucleotide sequences among 11 species (tiger, wolf, leopard, markhor, wild camel, olive baboon, gorilla, fox, yak, cheetah and human) with 13 samples of wild species while remaining were obtained from databases. Evolutionary analyses were conducted by using MEGA7. Figure 1 shows the relationship among all species of wild, domestic and other species taken from databases. This cladogram represents that each of the breed of goat is arising from the common node bears negligible mutation in the study and thus is placed in the same clade. All the sheep species showed more mutational events than goat. The minimum value for percentage mutational event in this study was found to be of Kamori goat breed with teddy that is 0% whereas the maximum value was found to be of camel 12.96 with horse.
Application of Bioinformatics Tools
Prot Param tool
Prot Param is an online bioinformatics tool which is used for observing the physiological features of protein. These properties help in better understanding the role of proteomics due to which species could have altered pathways hindering the metabolic activities. Wild type of The “Extinction Coefficient” explains the amount of light that is absorbed by the protein. The half-life of a protein specifies the time when the protein is degraded into half after its synthesis. The “Aliphatic Index” indicates the volume occupied by the aliphatic side chains. Instability index describes stability of proteins in test tube, if value is above 40 than protein is unstable and results indicated that wild type is highly stable whereas mutant protein is unstable in our study. This stability of proteins might have an effect on behavioral characteristics of wild type as compared to docile animals. “GRAVY” indicates the hydrophobicity of the protein; high positive score indicates high hydrophobicity, while our results showed negative values.
Motif Scan tool
It (http://myhits.isb-sib.ch/cgi-bin/motif_scan) is another bioinformatics tool of great importance that gives information about different motifs present in the particular sequence of the protein. It allows the alignment and match scoring of the sequence with its database thus providing interpretation of the matched scores. Results in domestic species showed that LDL receptor class B is present in wild type of domestic species whereas mutants have Nebulin repeat profile in common. Results in wild species showed that wild type, Mutant 2,3,4 and 5 has no domain in it whereas one of the mutants i.e. M-1 has bacterial domain.
Prosite scan tool
It scans proteins for matches against the Prosite database of motifs as well as consensus patterns. Nascent protein undergoes post-translational modifications and becomes functional. Thus, it is necessary to study those modifications, this tool also provides the information about the post translational modifications.
Post translational modifications of the wild and mutant protein are shown in Table 5 and 6. Any one of the amino acids mentioned in this bracket “[ ]” may be present at this position, while an amino acid within“{}”shows that there can be any amino acid at that site except for the one indicated within the braces. Similarly, parenthesis “( )” shows the number of amino acids that should be present at a particular site and “x” shows any of the amino acids [12,13].
Tables & Figures
Mutations were read after aligning the sequences from NCBI Blast2Sequece. Coding DNA sequence file of wild and docile species was made which helped in counting the base pairs at the position where polymorphisms were present [12]. Polymorphisms in two samples of Mungli breed were observed and it was found that both of them had deletion at 956 of coding position but this polymorphism had no effect on amino acid change. Glutamine protein was being converted into glutamine even after this polymorphism. Thus, it was regarded as synonymous mutation. Lehri goat had polymorphism at the same position. At c.956 position and 1063, Adenine was being converted into guanine (in both the cases), due to which glutamine changed into arginine (in the first polymorphism) and isoleucine changed into valine (in the second polymorphism). Angora goat, Kamori goat 1, Kamori goat 2, Teddy goat 1 and Teddy goat 2 had polymorphism at c.956 where ‘A’ was being changed into ‘G’ which affected the protein product due to change in amino acid sequence. Glutamine was changed into arginine. These polymorphisms can be regarded as non-synonymous ones in coding region of exon no. 8 [14]. Similar mutations were found in a study of a point mutation within a codon in exon 8 of the human MAOA leading to a premature termination of protein which is associated with uncertain mental retardation as well as impulsive aggression, arson, attempted rape and exhibitionism in male members of a Dutch family [9].
Phylogenetic tree shows that Rawal tiger and wild leopard are closely related to one another on the basis of MAOA gene sequence. 0.03% mutational events are present in wild leopard. Markhore is the next related species present nearer to them with 2.7% (1.39+1.31) mutational events in it. Wild wolf and Mohini tigress have been originated from the same node and thus are sister groups with 2.09% mutational events. Wild Bactrian camel and Olive baboon has 2.80% mutation. This tree has been constructed based on the sequence of one gene rather than the whole genome. Still, these results could help guide phylogenetic analysis on the basis of whole genome given that MAOA the candidate gene in this study plays an important role in influencing behavior [11].
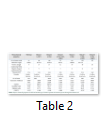
Prot Param tool was applied for analyzing the physio-chemical properties in domestic mutants. Wild type of MAOA Protein has least number of amino acids and Mutant 5 has the maximum molecular weight. Physio-chemical properties in this variant have been greatly affected. The “Extinction Coefficient” value for wild type and M6 was found to be same. The half-life for wild type, M3, M4 and M5 was same i.e. 1.2 hrs. Instability index indicated that wild type is highly stable whereas mutant protein is unstable [15]. The “Aliphatic Index” value for wild type, M1, M2 and M4 is same. “GRAVY”, results showed negative values. Motif scan tool was used [16]. Nebulin repeat profile was found in wild type, M1, M2, M3, M4 and M5 whereas M6 had protein prenyl transferase alpha subunit repeat profile.
Sequence diversity analysis was performed on wild species. Four species (Tiger, Leopard, Markhore and wild wolf) of different species (Tiger Rawal, Sam tiger, Mohini tigress, Wild leopard, wolf and Markhor) were analyzed among wild animals. Polymorphisms in three breeds (Rawal tiger, Sam tiger and Mohini tigress) of one species (Tiger) were observed. The results showed that Rawal tiger and wild leopard had a polymorphism at coding position of c. 2530 in exon no. 15 in which the base changed into Thymine. This one base change had an adverse effect on protein product of both (Rawal tiger and Leopard) the species as reading frame was shifted [15]. Tiger Sam and Mohini tigress had a polymorphism at c. 2528 position in which the base changed into thymine and Glutamine amino acid was converted into serine [12]. A study was conducted in which three polymorphisms (rs909525,rs6323 and rs2064070) of the MAOA gene were found to be highly associated with aggression related traits in suicidal males, while the single nucleotide polymorphism(SNP) rs6323 was also found associated with anger in females [6]. These variations lead to a clue that this single gene, though present in both wild and docile species, is present in diversified form. Sequence diversity can be clearly observed within each wild and docile species.
Prot Param tool was applied for analyzing the physio-chemical properties in wild species mutants as well. Wild type of MAOA Protein has least number of amino acids i.e. 52. Mutant 1 has the maximum weight. The “Extinction Coefficient had the least value in wild type whereas Mutant 2 had the highest value [15]. The half-life for wild type, M3, M4 and M5 was same i.e. 1 hr. Instability index indicates that wild type is highly stable whereas mutant protein is unstable [16]. The “Aliphatic Index” value for wild type, M3 is same. “GRAVY” results showed positive value in M4 only. Extra post translational modification amidation site was found in wild species and this was absent in domestic in species.
CDS position c.956 was observed as common variant in docile species, while none of the position were found common among wild animals. Phylogenetic tree was constructed on the basis of these observed variants to have an insight of existing taxonomy of these animals. Observed variants in MAOA gene might be responsible for behavioral differences between wild and docile species and may be helpful in designing new studies for confirmation and to ascertain the association of these altered loci in the larger purview of systems biology.
The authors declare that there is no conflict of interest regarding the publication of this paper.
- Heath MJ, Hen R. Serotonin receptors: Genetic insights into serotonin function.Journal of Consumer Behavior, (1995); 5(9): 997-9.
- Daubert EA, Condron BG. Serotonin: a regulator of neuronal morphology and circuitry. Trends in Neurosciene, (2010); 33(9): 424-34.
- Wellman CL, Camp M, Jones VM, MacPherson KP, Ihne J, Fitzgerald P, et al. Convergent effects of mouse Pet-1 deletion and human PET-1 variation on amygdala fear and threat processing. Experimental Neurology, (2013); 250: 260-9.
- Beaver KM, Nedelec JL, Wilde M, Lippoff C, Jackson D. Examining the association between MAOA genotype and incarceration, anger and hostility: The moderating influences of risk and protective factors. Journal of Research in Personality, (2011); 45(3): 279-84.
- Adamczyk K, Pokorska J, Makulska J, Earley B, Mazurek M. Genetic analysis and evaluation of behavioural traits in cattle. Science, (2013); 154(1): 1-12.
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, et al. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron, (2003); 37(2): 233-47.
- Brunner HG. MAOA deficiency and abnormal behaviour: perspectives on an association. Nature, (1996); 194: 155-64.
- Zalsman G, Patya M, Frisch A, Ofek H, Schapir L, Blum I, et al. Association of polymorphisms of the serotonergic pathways with clinical traits of impulsive-aggression and suicidality in adolescents: a multi-center study. Springer, (2011); 12(1): 33-41.
- McDermott R, Tingley D, Cowden J, Frazzetto G, Johnson DD. Monoamine oxidase A gene (MAOA) predicts behavioral aggression following provocation. Proceedings in National Academy of Sciences, (2009); 106(7): 2118-23.
- Wendland J, Hampe M, Newman T, Syagailo Y, Meyer J, Schempp W, et al. Structural variation of the monoamine oxidase A gene promoter repeat polymorphism in nonhuman primates. Genes, Brain and Behavior, (2006); 5(1): 40-5.
- Tamura K. and Nei M. Estimation of number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and evolution, (1993); 10: 512-526.
- Cases O, Seif I, Grimsby J, Gaspar P, Chen K, Pournin S, et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in mice lacking MAOA. Science, (1995); 268(5218): 1763.
- Haskell MJ, Simm G, Turner SP. Genetic selection for temperament traits in dairy and beef cattle. Frontiers in genetics, (2014); 5: 368.
- Shah SS, Mohyuddin A, Colonna V, Mehdi SQ, Ayub Q. Monoamine Oxidase A gene polymorphisms and self reported aggressive behaviour in a Pakistani ethnic group. Journal of Pakistan Medical Association, (2015); 65(8): 818-24.
- Beevers L. Post-Translational Modifications. In: Nucleic Acids and Proteins in Plants i. Springer, (1982); 136-68.
- Dorfman HM, Meyer-Lindenberg A, Buckholtz JW. Neurobiological mechanisms for impulsive-aggression: the role of MAOA. Neuroscience of Aggression: Springer, (2014) p. 297-313.
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. European Journal of Biochemistry, (2001); 268(10): 2764-2772.
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI ,Lanczycki CJ.CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Research, (2013); 41: 348-352.
- Idicula-Thomas S, Balaji PV. Understanding the relationship between the primarystructure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Science: A Publication of the Protein Society, (2005); 14: 582-92.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0