Full Length Research Article
Rapid RNA Extraction from Eucalyptus tree and its down processing for cloning of dehydrin genes
Ghulam Zahara Jahangir1, 2, Shagufta Naz*,2, Muhammad Idrees Khan1, 3, Muhammad Zafar Saleem1
Adv. life sci., vol. 5, no. 4, pp. 185-191, August 2018
*- Corresponding Author: Shagufta Naz (Email: snaz31@gmail.com)
Authors' Affiliations
2- Departments of Biotechnology, Lahore College for Women University, Lahore, Pakistan
3- Hazara University, Mansahra, Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: RNA extraction from tree species like Eucalyptus is very tedious and difficult task. In this study a very short and efficient method of RNA extraction from Eucalyptus has been described.
Methods: A very short and efficient protocol of two steps RNA extraction has been optimized for obtaining high quality and pure RNA from different tissue types of Eucalyptus tree. In first step whole nucleic acid was extracted from plant tissues and in second step RNA was purified from nucleic acid mixture. The newly optimized rapid CTAB RNA extraction method was compared with trizol extraction method for efficiency and quality of extracted RNA.
Results: The newly optimized rapid CTAB RNA extraction method was found highly efficient and suitable over the trizol method. Three Eucalyptus dehydrin genes the dehydrin-10 (DHN-10), dehydrin-1 (DHN-1), and dehydrin-2 (DHN-2) were successfully amplified, TA cloned into pTZ57/RT vector, and transformed into Top10F’ strain of E.coli. These three Eucalyptus dehydrin genes have been reported for conferring abiotic stress tolerance to the Eucalyptus plant yet have not been reported to be cloned. These cloned genes would be further manipulated for developing abiotic stress tolerance in plants of interest.
Conclusion: Rapid CTAB RNA extraction method is a brief and reproducible methodology for a hard job of RNA extraction from tree plants with high phenolic contents.
Keywords: Dehydrin proteins, Eucalyptus abiotic stress tolerance proteins, cloning of dehydrin genes, DHN-1, DHN-2, DHN-10
Although a sufficient magnitude of work has been done for RNA extraction from the woody plants, forest trees and perennial plants. Even then RNA extraction from forest tree is a big challenge for university labs of low-income countries like Pakistan. It is not possible for most of our labs, at university level, to follow the published protocols cent percent. The main hindrance is the lack of expensive equipment like tissue homogenizer, refrigerated ultracentrifuges etc, which are used for successful RNA extraction in already published methods [1]. This rapid RNA extraction protocol is designed for obtaining purified RNA from Eucalyptus and is simplification of work of Gambino et al [1]. This is a very successful and short protocol of less than three hours. This method is also compared with conventional trizol RNA extraction method and was not found successful for Eucalyptus RNA. Another plus point of the presented method is that it yields total nucleic acid first from which both RNA as well as DNA can be purified and used separately.
Eucalyptus is an important and main tree for commercial forestry and is being extensively studied due to its highly demanded wood quality and fast growth [2]. The presented study is impressed by the higher tolerance of Eucalyptus tree to the environmental abiotic stresses like low temperatures, drought and excessive salts. Three dehydrin proteins (of 10, 20 and 30 KDa) of Eucalyptus globulus have been reported to be associated with the cold tolerance of the Eucalyptus plant [3]. Keller et al proposed a strong relation between dehydrin 1 (DHN1) of Eucalyptus gunnii and cold tolerance [4]. The DHN-2 gene of E. gunnii has also linked with its superior cold-acclimation [5]. Dehydrin proteins are reported to make important group of Late Embryogenesis Abundant proteins, the LEA D-11 family. The dehydrin proteins are described to be the most conspicuous group of water soluble proteins that are inducible by a dehydration stress. The induction of these proteins has been linked with cold acclimation, with drought and salinity stress, abscisic acid responses and embryo development in over 100 independent studies [6]. The dehydrin proteins have been linked with tolerance against abiotic stresses [3]. The genetic manipulations of Eucalyptus dehydrin genes for developing abiotic stress tolerance in important commercial crops is one of the less studied aspect. Therefore the present study was focused on the isolation of the abiotic stress tolerance genes from Pakistani native Eucalyptus species and their TA cloning for further manipulations of interest.
Rapid RNA Extraction:
Preparation of Rapid RNA Extraction Buffer: 100ml of Rapid RNA Extraction buffer was prepared having following final concentrations: 2% CTAB, 2.5% PVP-40, 2M NaCl, 100mM Tris-HCl (pH 8.0), 25mM EDTA (pH 8.0). Chloroform-isoamyl alcohol mixture was used in the ratio of 24 and 1 respectively (v/v). 4M lithium chloride (LiCl) was prepared and added to supernatant for final concentration of mixture not less than 3M.
Protocol: The Rapid RNA Extraction Buffer was heated at 65ºC for 20 minutes in water bath. 100mg plant sample (hard bark, woody green twig tissue (green), fleshy green twig tissue, green twig near apices, older hard leaves, young soft leaves, young leaf primordia, and young floral buds) was grinded in liquid nitrogen in previously autoclaved plus DEPC treated pestle mortar and shifted to 1.5 ml eppendorf tube. Then 600 μl of hot (65ºC) Rapid RNA Extraction Buffer was added and the tubes were vortexed briefly. Then incubated the tubes at 65ºC (water bath) for 20 minutes. After incubation 600 μl of Chloroform: Isoamylalcohol (24:01) was added and shacked with hand vigorously. Centrifuged at 12000 RPM for 15 minutes at room temperature. Supernatant was collected to the new tubes and added LiCl for final concentration of not less than 3M. For this purpose the 4M LiCl was taken according to the supernatant volume like 375 μl of 4M LiCl was added into 500 μl supernatant. The mixture was mixed gently and incubated on ice for 30 minutes. Tubes were centrifuged at 14000 RPM for 20 minutes. Collected the supernatant into new tube and isopropanol was added in equal volume. Centrifuged at 14000 RPM for 20 minutes and supernatant was discarded. The pellet was washed with 500 μl of 76% ethanol at 14000 RPM for 05 minutes. Air dried the pellet for 8 minutes and re-suspended in 60 μl of sterilized water of molecular grade. After resuspension in water, the tubes were immediately put on ice or at -20ºC in refrigerator. In the next step RNA was purified from the extracted nucleic acid mixture. Before the purification of RNA from the extracted nucleic acid, 5 μl of the nucleic acid suspension was loaded on 1% agarose gel in 1x TAE Buffer. The voltage was set at 75 volts and run for 40 minutes in Mini Gel tank (5 volts/cm). The gel was observed under UV illuminator it showed DNA as well as three bands of RNA. Half of the mixture was used as DNA for amplification of full-length dehydrin genes. The remaining half mixture was processed for purification of RNA.
RNA purification:
50 μl of Trizol reagent was added to 50 μl of nucleic acid suspension and put at 37ºC for 5 minutes. Then centrifuged at 3000 RPM for 10 minutes. Collected supernatant to new tube and added 100 μl of chloroform. Vortexed for few seconds and again centrifuged at 3000 RPM for 10 minutes. The upper phase was collected into new tube and 500 μl of isopropanol was added. Mixed and centrifuged at 13000 RPM for 20 minutes at 4ºC. Discarded the supernatant and pellet was washed with 75% ethanol at 13000 RPM for 05 minutes at 4ºC. After slight air drying the pellet was resuspended in 30 μl of sterilized and DEPC treated water of molecular grade. 5 μl of RNA suspension was loaded on 2% agarose gel in 1x TAE Buffer. The voltage was set at 75 volts and run for 50 minutes in Mini Gel tank (5 volts/cm). The gel was observed under UV illuminator it showed only three bands of RNA and no DNA was visible.
RNA Extraction with Trizol:
Crushed 100 to 200 mg of Eucalyptus sample in a pestle mortar properly sterilized and previously treated with DEPC. A paste of sample was made by further crushing in 1.0 ml of Trizol (Invitrogen Cat No. 15596-018). Added 200 μl Phenol (Riedel-de Haen UN-No: 1671): Chloroform (Sigma Aldrich CAS-No. 67-66-3): Isoamylalcohol (Sigma Aldrich CAS-No. 123-51-3) in ratio of 25:24:01 and kept at room temperature for 5 minutes. Centrifuged the mixture at 12000 RPM for 15 minutes at 2-8ºC. Shifted to ice and collected aqueous phase into clean tubes and again put on ice. Then added 500 μl isopropanol and again put on ice for 30 minutes. After incubation, samples were centrifuged at 12000 RPM for 15 minutes at 2-8ᵒC before carefully decanting the supernatant and addition of 500 μl of 75% ethanol to the pellet. Finally, the dissolved solution was centrifuged for 10 minutes at 2-8ºC. The resultant RNA pellet was resuspended in DEPC treated injection water as a final product.
cDNA Synthesis:
cDNA was synthesized on the same day of RNA extraction. For 20 μl cDNA reaction the RNA was 10 μl; 1.0 μl of 100 pmol Gene specific reverse primer; 1.0 μl 10mM dNTPs mixture; and 1.0 μl of RNase inhibitor was used. The mixture was spun briefly and put at 65ᵒC in thermalcycler and immediately put on ice after for 5 minutes. Then added 4.0 μl of 5x cDNA Buffer (Invitrogen Cat no. 28025-013); 2.0 μl of DTT (Invitrogen Cat no. 28025-013); 1.0 μl of MMLV (Invitrogen Cat no. 28025-013). The reaction was run at 37ᵒC for 50 minutes and 70ᵒC for 15 minutes.
Amplification of dehydrin 1 (DHN-1), 2 (DHN-2) and 10 (DHN-10) genes from DNA and cDNA:
The three dehydrin genes, DHN-10, DHN-1, and DHN-2, were amplified from non-purified nucleic acid (DNA) extracted in the above method. The amplification was achieved with approximately 200 ng genomic DNA template, 0.2 mM dNTPs Mix (Promega# U1515), I X Taq Buffer, 2.5mM MgCl2 and 1.5U Taq polymerase (ThermoScientific # EP0402). Raised final reaction volume with PCR grade water up to 50μl. On Kyratec thermal cycler (SC300G) initial denaturation attained at 95ᵒC for 5 minutes, then 35 repeats of cycling conditions at 95ᵒC for 45 seconds, 54ᵒC for 45 seconds, 72ᵒC for 1 minute with final extension at 72ᵒC for 7 minutes. Later for amplification from plasmid DNA all conditions were same except the concentration of plasmid template, 30 to 50 ng of which was enough for good product yield. Naz et al., reported the primers for the full-length amplification of DHN-10, DHN-1, and DHN-2 genes that were used during the present study. For the amplification of protein coding sequences (exons only) of the said three dehydrins genes primers were designed on Primer3 software using published sequences of DHN-10, DHN-1, and DHN-2 under NCBI accession numbers JN052210.1, JN052208.1, and JN052209.1 respectively.. Sequences of these primer pairs have been given in the Table-1.
Cloning of dehydrin 1 (DHN-1), 2 (DHN-2) and 10 (DHN-10) genes:
The dehydrin 1 and dehydrin 10 genes that were amplified from RNA through cDNA were TA cloned into pTZ57/RT TA cloning vector of Thermo Scientific
InsTAclone PCR Cloning Kit (#K1213, #K1214) followed by the recommended protocol. The 5 μl of the resulting construct was transformed into 100μl of previously prepared competent cells of Top10F’ strain of E.coli through heat-shock method (putting construct-cell mixture on ice for 30 minutes, then a brief heat shock of 2 minutes at 42ºC, and immediately retuning back to ice for 3 minutes). After transformation 1ml of Luria’s Broth (LB) medium was added to the construct-cell mixture and transferred the mixture into a 15ml culture tube. The tube was put on shaking incubator at 37ᵒC. After one hour of constant shaking, 50μl to 100 μl of the culture was spread on LB Agar Plates having selection of 12.5mg/ml tetracycline and 100μg/ml Ampicillin drugs. The plates were put inverted in incubator at 37ᵒC overnight. For the screening of true transformants having properly ligated insert, through blue-white colony selection, X-Gal solution of 20mg/ml concentration (Thermo Scientific #0941) and 100 mM IPTG solution (Thermo Scientific #1171) were also added to the plates at the rate of 1μl/ml of medium. Single white colonies were picked and inoculated into 10ml LB broth medium having 12.5mg/ml tetracycline and 100μg/ml Ampicillin drugs and put on shaking incubator at 37ᵒC overnight. The 6ml of the overnight grown culture from single transformed colony was processed for plasmid isolation using FavorPrep TM Plasmid DNA Extraction Mini Kit (# FAPDE 100, 300) following the recommendations of the kit.
Confirmation of clones for containing Eucalyptus dehydrin genes: The plasmids purified from positive clones were processed through restriction digestion and PCR amplification with gene specific primers. For restriction digestion reaction of 20μl, added 1X Buffer, 5Units from EcoR1 and/or BamH1 restriction Enzyme(s), 1.0 μg plasmid DNA (10μl) and final volume achieved with injection water. The reaction tubes were put at 37ᵒC in an incubator for 2 hours and then checked on 1% agarose gel made in 1X TAE electrophoresis buffer at the speed of 5volts/cm. The conditions for PCR amplification of dehydrin genes from pTZ57R/T plasmid have been described above.
RNA Extraction
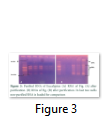
The two methods of RNA extraction, the conventional Trizol method and modified Rapid CTAB method, were compared and later found superior over former for efficiency, quality of RNA, simplicity, and for time saving properties. The quality of RNA obtained through rapid method is highly superior over conventional Trizol method. The RNA obtained at the end of Trizol method needs further purification (Fig-1 A & B). The extra step of purification requires more trizol and time. Furthermore, RNA could not be recovered from all tissue types of Eucalyptus tree used for RNA extraction in trizol method. Like green twigs, fleshy ends of green twigs that bear florescence and older leaves did not obtain RNA but leaf primordial and floral buds only and bands of RNA were also slightly sheared. On the contrary the quality of RNA as well as DNA obtained through Rapid CTAB method was excellent and self-explanatory (Fig-2 A & B). Distinct and compact bands for all three types of RNA were observed along with upper most sharp band of DNA. Furthermore, the RNA obtained at the end of Rapid CTAB method is highly pure (Fig-3 A & B).
Amplification of Eucalyptus Dehydrin 10, 1 and 2 genes:
The Eucalyptus Dehydrin-10, 1 and 2 genes were amplified from extracted DNA (nucleic acid mixture of DNA and RNA without any further purification of DNA). Full genes for all three dehydrins were observed and estimated at sizes of 525bp, 959bp and 1171bp for DHN-10, DHN-1, and DHN-2, respectively (Fig-4). To get the protein codng region of genes, all three dehydrin genes were also amplified from cDNA which was raised from purified RNA. The PCR products amplified through cDNA were estimated to be of sizes comparative to the reported sizes of 617bp, 777bp, and 297bp for DHN-1, DHN-2, and DHN-10 respectively (Fig.5 A & B).
Cloning of Eucalyptus Dehydrin Genes:
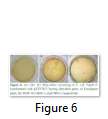
The whole gene as well as the coding sequences of Dehydrin-10, 1 and 2 genes were TA cloned into the pTZ57R/T vector. The ligation products, the constructs of pTZ57R/T having DHN-1, DHN-2, and DHN-10 separately, were transformed into E.coli Top10F’ strain. The attempt went successful and the clones were evaluated for successful ligation of inserts. Several white colonies (Fig.6) were randomly selected for growth in broth medium and then for plasmid prep. The beautiful bands of ligated pTZ57R/T plasmid were observed on agarose gel (Fig.7). From these plasmids DHN-10 gene (525 bp) was amplified using gene specific primers and single bands were observed at the position of slightly above the 500bp band of 100bp DNA ladder (Fig.8). Fig.9 shows double restriction of this plasmid with EcoR1 and BamH1 enzymes. The released insert along with 40 bases of the vector (total of 565bp) was visible at approximate size of last band of Lambda HindIII DNA marker (of 0.56 Kbp) and the empty linearized pTZ57R/T vector was visible at size larger than the 2.32 Kbp of same marker (of approximate size of 2847bp). The pTZ57R/T vector having Eucalyptus dehydrin 1 and 2 genes, digested with single enzyme, appeared on agarose gel at position of size slightly higher than 3000bp detects successful integration of inserts (Fig.10). Originally empty linearized pTZ57R/T vector is of 2887bp and its band should appear at position of size slightly lower than 3000bp (below the top most band of 100bp DNA marker).
Tables & Figures
Gambino et al., have reported an efficient method of RNA from different tissues of woody plants [1]. They have reported to yield RNA in three hours but their method uses high speed refrigerated centrifuges. This reported work is the modification of their work which yields excellent quality pure RNA without high speed refrigerated centrifuge. The simplifications are needed because most of the college and university labs of low income countries like Pakistan that are engaged with biological research are lacking expensive equipment like high speed refrigerated centrifuges, tissue homogenizing machines, and RNA extraction machines etc. The presented rapid CTAB RNA extraction method is an easy and economic extraction protocol. It uses one extraction buffer with single incubation at 65ᵒC and centrifugation at 14000rcf at room temperature; whereas the RNA extraction method of Gambino et al uses two extraction buffers, two incubations at 65ᵒC, and high speed refrigerated centrifuge for centrifugation at 21000g. Hence, the presented rapid CTAB RNA extraction protocol is more economical and time saving than that of the Gambino et al., [1].
Naz et al., have characterized three Eucalyptus Dehydrin genes, the dehydrin 10, dehydrin 1, and dehydrin 2, along with other Eucalyptus genes that confer abiotic stress tolerance to the Eucalyptus plant [7]. They have characterized these genes from different species of Eucalyptus that are commonly grown in Pakistan like E. globulus, E. camaldulensis, E. gunnii and E. tereticornis. Their work reports amplified PCR products for DHN-10, DHN-1, and DHN-2 from Eucalyptus DNA at 525bp, 959bp and 1171bp sizes respectively which are in close accordance of this work. Fernandez et al have also reported that coding region of DHN-1 gene is of 617bp, DHN-2 of 777bp, and DHN-10 of 297bp [3].
Naz et al., have cloned the dehydrin 2 gene of Eucalyptus camaldulensis into TA cloning vector (pCR 2.1 cloning vector) [7]. None of other Eucalyptus dehydrin gene has been cloned to form some construct. However a considerable mass of work has been published upon the protein study of dehydrin genes. Some researchers have identified and cloned different dehydrin genes from various plant species like Yao et al have identified and cloned dehydrin genes from Brassica napus and Brassica juncea as a novel subclass of late embryogenesis-abundant protein [8]. Yakubov et al., have cloned a full-length cDNA with dehydrin-like identification from a Pistacia vera [9]. Yang et al have identified and cloned a novel dehydrin from Stipa purpurea (SK3 type) [10].
The presented work have found as an efficient and very brief method of whole nucleic acid extraction and successful purification of RNA from it for a woody tree species of Eucalyptus. Three dehydrin genes of Eucalyptus species, the dehydrin 10, 1 and 2, have been amplified from Eucalyptus DNA as well as RNA are also cloned into TA cloning vector. The cloning of three dehydrin genes of Eucalyptus is a new addition in the existing knowledge.
Conflict of interest
Authors declare that there is no conflict of interest for publishing this study.
Acknowledgement
The Plant Biotechnology Lab in the Department of Biotechnology, Lahore College for Women University provided the enzymes, molecular reagents, primers and consumables used during the present study. All of the experiments were designed and were executed at Agri-Biotechnology Lab (CAMB) using the research facilities of Centre for Applied Molecular Biology, University of the Punjab, Lahore. We are very thankful to kind cooperation of Proteomics Lab CAMB and DNA Core Facility of CAMB especially Mr. Muhammad Usman and Mr. Muhammad Akram for their generous help. We are also very thankful to the students of Agri-Biotechnology Lab CAMB especially Ms. Qurat-ul-Ain and Mr. Afzaal Gillani.
- Gambino G, Perrone I, Gribaudo1. A Rapid and Effective Method for RNA Extraction from Different Tissues of Grapevine and Other Woody Plants. Phytochemical Analysis, (2008); 19: 520–525.
- Girijashankar V. Genetic transformation of eucalyptus. Physiology and molecular biology of plants : an international journal of functional plant biology, (2011); 17(1): 9-23.
- Fernandez M, Águila SV, Arora R, Chen K. Isolation and characterization of three cold acclimation-responsive dehydrin genes from Eucalyptus globulus. Tree Genetics & Genomes, (2012); 8:149–162.
- Keller G, Marchal T, SanClemente H, Navarro M, Ladouce N, et al. Development and functional annotation of an 11,303-EST collection from Eucalyptus for studies of cold stress. Tree Genet Genomes, (2009); 5:317–327.
- Navarro M, Ayax C, Martinez Y, Laur J, Kayal WE, et al. Two EguCBF1 genes overexpressed in Eucalyptus display a different impact on stress tolerance and plant development. Plant Biotechnology Journal, (2010); 9:50–63.
- Porcel R, Azco R, Ruiz-Lozano JM. Evaluation of the role of genes encoding for dehydrin proteins (LEA D-11) during drought stress in arbuscular mycorrhizal Glycine max and Lactuca sativa plants. Journal of Experimental Botany, (2005); 56 (417): 1933–1942.
- Naz S, Kausar H, Saleem F, Zafarullah A. Characterization of abiotic stress genes from different species of eucalyptus. Pakistan Journal of Botany, (2015); 47(4): 1217-1223.
- Yao K, Lockhart KM, Kalanack JJ. Cloning of dehydrin coding sequences from Brassica juncea and Brassica napus and their low temperature-inducible expression in germinating seeds. Plant Physiology and Biochemistry, (2005); 43(1):83-9.
- Yakubov B, Barazani O, Shachack A, Rowland LJ, Shoseyov O, et al. Cloning and expression of a dehydrin-like protein from Pistacia vera L. Trees, (2005); 19: 224–230
- Yang Y, Sun X, Yang S, Li X, Yang Y. Molecular cloning and characterization of a novel SK3-type dehydrin gene from Stipa purpurea. Biochemical and Biophysical Research Communications, (2014); 448:145–150.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0