Full Length Research Article
First record of Cantharellus minor from Vietnam with identification support from a combination of nrLSU and nrSSU phylogenetic analysis
Thuan Duc Lao*, Nghia Trong K, Tai Van Ngo, Nguyen Binh Truong, Luyen Tien Vu, Thuy Ai Huyen Le
Adv. life sci., vol. 6, no. 3, pp. 125-130, May 2019
*- Corresponding Author: Thuan Duc Lao (Email: thuan.ld@ou.edu.vn)
Authors' Affiliations
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: A previously identified sample XC02, which was collected from a pine forest (Pinus kesiya Royle ex Gordon), in Xuan Tho Commune, Da Lat, Lam Dong Province, Vietnam, was identified as Cantharellus minor based on morphology and nrLSU phylogeny analysis. Sequence analysis of multiple genes are becoming more and more common for phylogenetic analysis of mushrooms.
Method: Total DNA was isolated from sample XC02. The primer NS1, NS4 were applied to amplify the target gene the nuclear ribosomal small subunit DNA (nrSSU). For phylogenetic analysis, individual and concatenated datasets (nrSSU and nrLSU-nrSSU) were constructed. Phylogenetic tree was constructed with MEGA 6.0 with a 1000 replicate bootstrap based on the neighbor joining, maximum likelihood, maximum parsimony method.
Results: A concatenated dataset containing a total of 14 sequences from Cantharellus, Craterellus (Cantharellaceae, Canthraellales) and Hydnum (Hydnaceae, Cantharellales) were constructed. For the specimen XC02, the phylogenies based on the first, second, and third datasets (nrLSU, nrSSU, and nrLSU-nrSSU) and the morphological analysis, reported in our previous study, strongly confirmed the identity of XC02 as Cantharellus minor.
Conclusion: The combination between the morphological analysis and phylogenetic analysis is confirmed as the best approach for the identification of Cantharellus and other mushroom species that we collected in the Central Highlands, Vietnam.
Keywords: nrLSU; Cantharellus, Cantharellus minor; nrSSU; nrLSU; phylogeny analysis; Vietnam
Cantharellus minor Peck (Mycobank# 179458), a popular edible and commercialized mushroom of the Cantharellus genus, annual Report on the New York State Museum of Natural History 23, 1872, was first identified in 1872 by Peck [1]. C. minor grows on soil, forming ectomycorrhizal association with Cedrus deodara, Quercus dilatata [2]. The distribution of C. minor was previously reported to be restricted only to the northern part of South America. However, current reports show that this species are present in the Western Ghats, Kerala, India, etc. [2]. Of note, C. minor has not been recorded in Vietnam. During our expedition to validate the fungal diversity in the Pine forest (Pinus kesiya Royle ex Gordon), a fast growing, has natural distribution in South-East Asia, ca. 1502m altitude, at Xuan Tho Commune, Da Lat City, Lam Dong Province, Vietnam, we collected sample XC02 which belong to the Cantharellus genus (Cantharellaceae) [1]. According to our previous report, the specimen was yellow, aromatic in flavor with smooth cylindrical stipe (1 – 2 mm in diameter and 20 – 50 mm in length). Pileus surface was smooth, scaleless, and yellowish (5 – 15 mm wide) with incurred, non-striated and wavy-like margin and infundibuliform yellowish to orange. Lamellae was similar to pileus in color, distant, decurrent without intervenose. Basidiospores were ovoid-ellipsoid with smooth surface (6 – 11.5 x 4 – 6.5 µm with each basidium containing 4 – 5 cornuted spores [1]. nrLSU phylogenetic analysis showed that XC02 formed a highly supported monophyletic group with referent sequence from Cantharellus minor, and separated this group from other referent taxa. Therefore, based on the above morphological description and molecular analysis of nrLSU, the specimen XC02 was identified as Cantharellus minor (Figure 1). Notably, this record was the first record of C. minor from Pine forest of Lam Dong, Vietnam.
Cantharellus and the closely related Craterellus genus, which can be recognized by their lack of division into cap, stipe and rudimentary, have recently been investigated using molecular phylogenetic analysis, including nrLSU, nrSSU, Rpb2 [2, 3]. Based on the phylogenetic analysis of nrLSU, nrSSU, mtSSU, and Rpb2 data, Cantharellus and Craterellus consistently formed monophyletic and sister groups thereby elucidating the phylogeny of species in Cantharellales order [4]. Moreover, the dataset of the publication is also the largest up to now on Genbank (NCBI).
In this current study, with the aim of strengthen the identification of XC02 as Cantharellus minor as well as and a wider scope to apply this data set onto the further analysis of samples that we collected in the Central Highlands, we conducted the molecular phylogenetic analysis of the combined set of nrLSU and nrSSU genes.
Fungus collection
The specimen XC02 was collected in a pine forest (Pinus kesiya Royle ex Gordon) near Xuan Tho Commune, Da Lat, Lam Dong Province, Vietnam (Figure 1). General pick up information: Height: 1.502 m; Humidity: 87%; Temperature: 20°C; Light intensity: 3.012 lux; Coordinates: 11o56'34.45" N, 108o28'33.56" E. In the laboratory, specimens were conditioned to room temperature, submerged in 1% Mercury (II) chloride for 5 – 10 minutes, dried at 60°C, and stored for further analysis.
DNA extraction, PCR assay and target gene sequencing
The phenol/Chloroform method (pH= 8) was applied to isolate genomic DNA. The fruit body was added to a lysis buffer (2.0% SDS, Tris-HCl pH 8.0, 150 mM NaCl, 10 mM EDTA, 0.1 mg/ml Proteinase K). During incubation at 65°C for overnight, it was mixed thoroughly by inverting the tube several times. Then, the supernatant was collected by centrifuged. 700 µL of PCI (Phenol/Chloroform/Isoamylalcohol with ratio of 25:24:1) solution was added and centrifuged. The upper solution was collected, precipitated with absolute ethanol, and washed with 70% ethanol. DNA concentration was identified by using OD260. Finally, isolated genomic DNA were stored in TE buffer at -20°C for further studies.
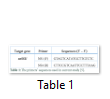
The primer pairs, used to amplify nrSSU region, are shown in Table 1. The final volume for PCR was 15 µL with a specified program: 1 cycle of 95°C for 5 min; 40 cycles of 95°C in 30 s, 42.2°C in 30 s, 72°C in 2 min; 1 cycle of 72°C in 5 min. 5 μl aliquots of amplification products were electrophoresed on a 2.0% agarose gel and visualized in a UV transilluminator. The amplified product was sequenced at Nam Khoa (Vietnam) Company with the same primers.
Taxa and sequences collection, DNA proofreading and phylogeny analysis
The data set of nrLSU, nrSSU were established by sequences downloaded from Genbank. The nrLSU, nrSSU were noted with strain, accession number, name of taxon and locality. The multiple gene data used in current study was established based on the combination of nrLSU and nrSSU data. Craterellus cornucopioides, Hydnum repandum and Craterellus tubaeformis were used as the outgroup for analysis. The amplified DNA sequences were proofread to remove ambiguous signals at both ends. Software, including SeaView 4.2.12, Chromas Lite 2.1.1, were used for proofreading. The phylogenetic tree was constructed, based on the neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods, by using Molecular Evolutionary Genetics Analysis (MEGA) version 6.0. Additionally, the best evolution model was predicted by using jModelTest [6].
The systematic concatenated nrLSU, nrSSU dataset
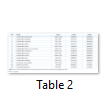
The nrLSU dataset included 59 sequences belonged to Cantharellus, and 1 sequence of Craterellus. The nrSSU dataset included 11 sequences belonged to Cantharellus, 2 sequence of Craterellus, and 1 sequence of Hydnum. The concatenated dataset was established based on individual data sets of nrLSU and nrSSU. As a result, the nrLSU-nrSSU dataset, which comprised of a total of 14 sequences representing 8 species of Cantharellus (Cantharellales, Cantharellaceae), 2 species of Craterellus (Cantharellales, Cantharellaceae), and 1 species of Hydnum (Cantharellales, Hydnaceae), was collected from Genbank (NCBI) and listed in Table 2.
nrSSU amplification and the nrSSU phylogeny analysis
In current study, extracted and purified DNA was amplified with NS1 and NS4 primers. Electrophoresis on 2.0% agarose gel showed a significant and unique band of 1,102 bp. The PCR product was sequenced. Sequencing signals of both strands were similar and good for reading (Figure 2). nrLSU amplification and alignment (the first dataset, containing 59 sequences) were reported in our previous study [3]. The evolution model that was most fit with the nrLSU dataset was Kimura 2-parameter. As a result, XC02 formed a highly supported monophyletic group with referent sequences from Cantharellus minor (Accession number: KF294625, KF294632) [1].
For the nrSSU dataset (the second dataset, containing 14 sequences and an XC02 sequence), phylogeny analysis was conducted based on the topology constructed by Maximum parsimony (MP), Neighbour Joining (NJ), and Maximum likelihood (ML) with 1000 bootstrap replication. The topology analysis showed that XC02 formed a monophyletic group with Clade 4 referent sequences. The XC02 sequence, within this clade, was strongly supported on the monophyletic group with referent sequence: Cantharellus minor (Accession: DQ898672), based on the observed bootstrap values of 98, 96, 97 (for ML, NJ, MP methods, respectively) (Figure 3).
The concatenated dataset of nrLSU-nrSSU analysis
The best-fit model of DNA evolution for the analyses, for the concatenated dataset (the third dataset), was obtained using the jModelTest2 [6]. The GTR + G model was the most-fit model employed across sites for nrLSU-nrSSU dataset. Phylogenetic analysis was presented in Fig. 4. The Cantharellus in the concatenated dataset again formed a strong monophyletic group and separated from the outgroup. All the species in our dataset formed 3 clades that were previously reported as the sub-generic groups of Cantharellus, including the subgenus Cantharellus (Clade 1), Cinnabarinus (Clade 3), Parvocantharellus (Clade 4). Notably, the subgenus Rubrinus (Clade 2) was not observed due to the missing nrSSU database in Genbank (NCBI). XC02 formed a monophyletic group in the subgenus Parvocantharellus, including Cantharellus appalachiensis and Cantharellus minor. Notably, within this clade, the XC02 formed a highly supported monophyletic group with referent sequence: Cantharellus minor with very high bootstrap values for ML, NJ, MP, respectively, and separated this group from the other referent taxon.
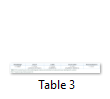
In the phylogenies based on the first, second, and third datasets (nrLSU, nrSSU, and nrLSU-nrSSU) and the morphological analysis, reported in our previous study, XC02 shared the same result as Cantharellus minor (Table 3). Therefore, the molecular analysis strengthened the identification of XC02 as Cantharellus minor. Moreover, the construction of the three datasets will facilitate further analyses of other samples collected in the Central Highlands.
Figures & Tables
The pine forest (Pinus kesiya Royle ex Gordon), ca. 1502 m altitude, at Xuan Tho Commune, Da Lat, Lam Dong Province, Vietnam is located in the Central Highlands, which is a unique area that is a hotspot mushroom biodiversity [7]. Therefore, the natural composition of mushroom species needs to be studied, developed and preserved. In the trip to validate the fungal diversity in the Central Highlands of Vietnam, the sample XC02, identified as C. minor based on the morphological analysis and nrLSU dataset phylogenetic analysis, was collected. C. minor is the native fungus to the Eastern North America [8]. Recently, C. minor has been reported to be discovered in evergreen forests in the Western Ghats, Kerala, India, Malaysia [9].
In previous study, the records of Cantharellus species from the northwestern Himalayas of India were reported [2]. In their report, they provided the key to identify all the recognized species based on the morphology and the molecular database of ITS and nrLSU sequence. Among those species, C. minor formed the monophyletic group with C. cinnabarius based on the nrLSU, and formed the group with C. elongatipes, C. appalachiensis, and C. cinnabarius based on the ITS. However, C. minor differed mainly based on pileus size and colour. Notably, although Cantharellus species were recorded from over the world, only few molecular databases are available. Therefore, in addition to its phylogenetic analysis to the study of Cantharellus taxanomy, the combination of the species having many sequences, such as ITS, nrLSU, nrSSU, etc. may enhance the higher phylogenetic analysis power. Therefore, in this study, to strengthen the identification of XC02 and toward a wider scope to apply this data set onto the further analyses of our samples, we reconstructed the phylogenetic trees using these sequences of nrLSU and nrSSU further analysis. Therefore, in addition to the first dataset (nrLSU), the second and third dataset (nrSSU, nrLSU-nrSSU) were created and analyzed by MEGA [10]. Both datasets showed the strongly supported species group subclades in all the dataset for phylogeny of Cantharellus. For the specimen XC02, as a result, there were no conflicts in the identification of evolution relationship between referent sequences in all dataset. The highly supported monophyletic group with referent C. minor was obtained with high bootstrap value (> 95), indicated that XC02 is closely related to C. minor. Phylogenetic analyses based on individual and concatenated datasets revealed clades with statistical and strongly support corresponding to morphological observation. Thus, XC2 was concluded as C. minor. Although Cantharellus species are recored and reported from all over the world, only few species have nrSSU sequences in public database (Genbank, NCBI), indicating the poor coverage of these species. However, in summary, combining the morphological and molecular data is the clearly the best approach to make the progress in the further studies of samples that we collected in the Central Highlands region. We have successfully applied the phylogenetic analyses based on individual (nrLSU, nrSSU) and concatenated datasets (nrLSU-nrSSU) to strengthen the identification of XC02 as Cantharellus minor. Additionally, the combination between the morphological analysis and phylogenetic analysis is confirmed as the best approach to apply this onto the further analysis of Cantharellus and other mushroom samples that we collected in the Central Highlands, Vietnam.
Acknowledgement
We express our special thanks to Faculty of Biology, Da Lat University and Faculty of Biotechnology, Ho Chi Minh City Open University for the genuine support throughout this research work.
The authors declare that there is no conflict of interest regarding the publication of this paper.
- Peck CH. Report of the Botanist (1869). Annual Report on the New York State Museum of Natural History. (1873); 23:27-135.
- Deepika K, Reddy MS, Upadhyay RC. New records of Cantharellus species from the northwestern Himalayas of India. Mycology. (2013); 4(4):205-220.
- Phan HH, Lao DT, Le HAT, Hoang QK, Truong BN, Ngo TLG. First record of Cantharellus minor in Vietnam. Journal of Biotechnology, (2017); 15(4): 669-673.
- Dahlman M, Danell E, Spatafora JW. Molecular systematics of Craterellus: cladistic analysis of nuclear LSU rDNA sequence data. Mycological research, (2000); 104(4): 388-394.
- Moncalvo JM, Nilsson RH, Koster B, Dunham SM, Bernauer T, Matheny PB, et al. The Cantharelloid clade: dealing with incongruent gene trees and phylogenetic reconstruction methods. Mycologia, (2006); 98(6): 937-948.
- Tibuhwa DD, Saviæ S, Tibell L, Kivaisi AK. Afrocantharellus gen. stat. nov. is part of a rich diversity of African Cantharellaceae. IMA fungus, (2012); 3(1): 25-38.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of bacteriology, (1990); 172(8): 4238-4246.
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature methods, (2012); 9(8): 772.
- Nguyen PDN, Tran DK. Impacts of ecological factors on the distribution of Amauroderma murrill genus in Central Highlands of Vietnam. Journal of Scientific and Engineering Research, (2017); 4(9): 238-243.
- Kuo M. Cantharellus minor. MushroomExpert.Com. (2006) Retrieved 2011-03-24.
- Deepika K, Reddy MS, Upadhyay RC. New records of Cantharellus species from the Northwestern Himalayas of India. Mycology, (2014); 4(4): 205-220.
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in bioinformatics, (2008); 9(4): 299-306
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0