Full Length Research Article
Arabidopsis Argininosuccinate Lyase and Argininosuccinate Synthase are important for resistance against Pseudomonas syringae
Shahbaz Anwar*, Muhammad Amjad Ali, Amjad Abbas, Krzysztof Wieczorek
Adv. life sci., vol. 7, no. 1, pp. 20-26, November 2019
*- Corresponding Author: Shahbaz Anwar (Email: shahbazanwarch@hotmail.com)
Authors' Affiliations
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Arginine is one of the important amino acids and is involved in a variety of plant responses against biotic and abiotic stresses. Two important genes Argininosuccinate Synthase (AS) and Argininosuccinate Lyase (AL) are associated with the production of arginine from ornithine. Here we report molecular characterization of Arabidopsis thaliana artificial micro RNA (amiRNA) mutants for the genes AL and AS in response to infection with Pseudomonas syringae DC3000.
Methods: Quantitative real time PCR (qRT-PCR) and Promoter::GUS plant lines were used for expression analysis. Artificial micro RNAs (amiRNA) mediated gene silencing was used to generate mutant plant lines. Flood inoculation technique was used for infection test essays with Pseudomonas syringae.
Results: Expression analysis of A. thaliana plants harboring promoter AS::GUS construct showed strong promoter activity upon P. syringae infection. Quantitative real time PCR (qRT-PCR) analysis showed that AL and AS expression was strongly induced upon infection with P. syringae. Infection essays for P. syringae showed enhanced susceptibility to virulent (Pto) as well as avirulent (∆avrPto/∆avrPtoB) strains of P. syringae. However, mutant plants infiltered with infiltration medium containing 1 mM L-arginine regain their resistance in comparison with wild type (Col-0) plants.
Conclusion: Our findings suggest that genes related to arginine metabolism play a key role in plant defenses during P. syringae infection on A. thaliana. This study revealed that proper functioning of arginine related genes is required to deploy defense response against P. syringae. Decrease in the expression of these genes improves conditions for the growth of pathogen.
Keywords: Arabidopsis thaliana; arginine; Argininosuccinate Lyase; Argininosuccinate Synthase; Gene silencing; Pseudomonas syringae
Plants are exposed to a number of infectious organisms, however successful disease progression depends upon specific molecular interactions between host and pathogens. Plants are equipped with specialized defense strategies deployed locally or systemically. Most of these defense reactions are associated with basal defense also called non-host resistance. This is the most prevalent form of resistance at species level [1,2]. Several cellular pathways are required to be activated for this type of resistance in plants e.g. induction of pathogen responsive genes, generation of reactive oxygen species, production of nitric oxide and callose deposition to reinforce the cell walls [3-6]. Different amino acids pathways are involved in plant-pathogen interactions as well as in developmental responses to several stress responses [7-9].
Nitric oxide (NO) is a free radical gas, which acts as signal during the induction of defense responses in plants [10,11]. In A. thaliana arginine is the main source of nitric oxide during plant-pathogen interactions [12]. Arginine is synthesized in plants using ornithine as precursor. This is two-step process including Argininosuccinate Synthase (AS) which converts citrulline and aspartate into argininosuccinate then Argininosuccinate Lyase (AL) further converts argininosuccinate into arginine and fumarate [14]. Decrease in arginine content in A. thaliana results in decreased NO activity [12].
Arginine plays a central role during biotic as well as abiotic stress. Arginine provide precursor for many signal molecules like nitric oxide and polyamines during stress responses. Polyamines are important molecules during cell proliferation and programmed cell death. Nitric oxide has direct role in plant defense responses. Arginine also serves as precursor for proline biosynthesis. Proline is an important osmolyte produced during stress responses.
Due to importance of arginine in plant NO synthesis and defense response, we report the molecular characterization of genes coding for two key enzymes Argininosuccinate Synthase (AS) and Argininosuccinate Lyase (AL) associated with the production of arginine amino acid. Our findings suggest that proper functioning of these two genes is required to interfere with the population growth of P. syringae on A. thaliana leaves.
Culture of Arabidopsis thaliana and Pseudomonas syringae
Wild type A. thaliana (Col-0) plants were used in this study. Seeds were planted in 2X2 inch plastic pots. Day length cycle was set at 16 h light followed by 8 h dark and at 25ºC in green house. These plants were used for transformation. A. thaliana -P. syringae interactions were studied in sterile culture on a Murashige and Skoog’s (MS) medium. Plates were incubated at cycle of 16 h light followed by 8 h dark and 25ºC. After 15 days seedlings were inoculated with P. syringae using seedling flood inoculation technique [16].
Plasmid construction and plant transformation
To clone promoter AS::GUS line, genomic DNA was extracted from A. thaliana using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), briefly ~ 418 bp fragment upstream from start codon of At4g24830 gene was amplified. Primers, pAS-Fr and pAS-Rv (Table 1) were used for promoter amplification. Sequences for the SgsI/AscI and HindIII restriction sites were included in primer design. Primers were designed using NCBI primer design tool (http://www.ncbi.nlm.nih.gov/tools/primerblast). Binary vector pMDC139 [17] was digested with SgsI/AscI-HindIII restriction enzymes. Amplified promoter sequence was cloned in binary vector.
Stable artificial micro RNAs (amiRNA) for AL and AS genes were constructed according to Schwab et al. [18]. Briefly sequences of four oligonucleotides for each AL and AS amiRNAs were identified. WMD Web Micro-RNA Designer was used to design oligonucleotides (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi). These oligonucleotides (Table 1) were used to amplify single amiRNA. Native A. thaliana miR319a precursor was used for site-directed mutagenesis. Modified pPZP3425 vector [19] was used to clone amplified amiRNAs. Presence of the inserts was confirmed by sequencing the binary vectors. Agrobacterium mediated plant transformation (floral dip technique) was used to insert binary vectors into the A. thaliana genome [20]. Screening of transformed plants was done on MS medium supplemented with kanamycin (50 µg mL-1). Homozygous T3 (third generation) lines were collected after segregation analysis. Silencing efficiencies of amiRNAs were tested in selected homozygous lines by qRT-PCR.
Infection test assays for Pseudomonas syringae
Flood inoculation technique was used to infect A. thaliana seedlings according to Ishiga et al. [16]. Briefly fifteen day-old seedlings were inoculated under sterile hood. Inoculum was consists of 40 mL bacterial suspension prepared in sterile distilled H2O containing 0.025% Silwet (L-77) referred as control. L-Arginine was added in 1mM concentration [12] in infiltration medium and was referred as treatment. We use virulent strain of P. syringae DC3000 (Pto) as well as avirulent strain of P. syringae DC 3000 (∆avrPto/∆avrPtoB). After 3 min of seedling submergence, bacterial inoculum was removed from plates by decanting. Petri plates were then sealed again and placed in the growth chamber. Internal bacterial population was determined after 1, 2 and 3 days post inoculation (dpi). Four seedlings were harvested from each plate which represents one biological replicate; three biological replicates from each plant line were taken for infection essays. Weight of one replicate was recorded and seedlings were surface sterilized three times with 70% ethanol. One washing was given with sterile dH2O. Samples were homogenized in 1.5 ml tubes and plated on LB medium containing the rifampicin (50µg/ml). Colony forming units (CFU) were counted from serial dilutions (10-2 to 10-6) and normalized against the weight of the sample.
Histochemical GUS assays
To study the expression patterns of AL and AS in A. thaliana leaves during P. syringae infection, GUS reporter gene was delivered under the control of native promoters for AS and ARGAH2 genes. Promoter::GUS lines were infected with P. syringae and GUS activity was determined. X-Gluc. (5-bromo-4 chloro-3-indolyl β-D-glucuronic acid) reagent was used for GUS staining assay. Five week old plants were inoculated as described above. One day post inoculation (dpi) rosette leaves were harvested and immersed in X-Gluc. solution. Petri dishes containing leaves in X-Gluc. solution were subjected to vacuum for 3 min and then incubated at 37ºC for four hours, after that these leaves were destained using 70% ethanol. Images of the leaves were taken with microscope (Axiovert 200M, Zeiss) containing a digital camera (Axiocam, Zeiss). Two pAS::GUS [15] and pARGAH2::GUS [21], T3 generation plant lines were studied for characterization of expression patterns.
Sample collection, RNA isolation and quantitative RT-PCR
Leaves were harvested for RNA isolation from A. thaliana plants. Samples were taken from plants infected with P. syringae at 1, 2 and 3 dpi. Harvested leaves were shock-frozen in liquid nitrogen. A. thaliana (Col-0) plants were used as control to compare amiRNA lines and non-infected plants of same age were used as control for P. syringae infected plants. Three independent biological replicates were used from each plant line to collect samples. Total RNA was extracted from A. thaliana leaves using RNase Plant Mini Kit (Qiagen, Germany). Quantity and quality of the RNA was analyzed using Agilent 2100 bio-analyzer (Agilent Technologies, USA). Random primers [oligo(dN)6] were used to reverse-transcribe RNA into cDNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Real-Time PCR System “ABI PRISM 7300 Sequence Detector” (Applied BioSystems, USA) was used to perform Quantitative RT-PCR. Program for PCR consists of 50oC for 2 min 95oC for 5 min and consecutive 40 cycles of 95oC for 15 s, 60oC for 30 s and 72oC for 1 min stages. 18S rRNA gene was used as internal references. Primer sequences are given in Table. 1. Fold changes were calculated using 2-ddCt method [22].
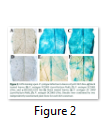
Native Promoter of argininosuccinate synthase and arginase 2 were cloned with GUS gene. Activities of pAS::GUS and pARGAH2::GUS constructs were studied in P. syringae infected and control A. thaliana leaves. Strong promoter activity was found in pAS::GUS lines upon infection with virulent strain of P. syringae (Pto) (Figure 2C) same was the case with avirulent strain (∆avrPto/∆avrPtoB) (Figure 2B). Promoter activity of pARGAH2::GUS was limited to few regions of leaves infected with Pto (Figure 2F) whereas ∆avrPto/∆avrPtoB infection resulted in a very week promoter activity (Figure 2E). Mock infiltrated control leaves of pAS::GUS and pARGAH2::GUS lines did not express any promoter activity (Figure 2A &D).
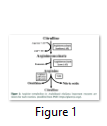
Expression of four genes playing key role in arginine metabolic pathway was determined by qRT-PCR during P. syringae infection on A. thaliana leaves. Two genes AS and AL associated with anabolic enzymes in arginine metabolism (Figure 1) were found significantly up-regulated at 1, 2 and 3 dpi. However, in case of virulent strain of DC3000 (Pto) gene expression was significantly higher at 2 and 3dpi as compared to 1dpi (Figure 3A). In case of ∆avrPto/∆avrPtoB infection expression was higher at initial time point and decreased at 2 and 3dpi (Figure 3B). Gene expression of ARGAH1 and ARGAH2, catabolic genes of arginine pathway (Figure 1) was not changed upon infection with both strains of P. syringae as compared to control samples of A. thaliana (Figure 3A & B).
To elucidate the involvement of enzymes in arginine metabolism, amiRNA-based gene silencing was done. Mutant plant lines for AS and AL were developed. Gene silencing by amiRNA was done because dominant null mutation may prove to be lethal for these genes. Homozygous T3 lines were selected by segregation analysis using kanamycin as described in the methods section. Expression of AS and AL in selected lines was determined using qRT-PCR. Mutant lines were selected based on their gene expression profiles. Two mutant lines as5-2 and as9-2 shows 60% decrease in gene expression for AS. In case of AL two mutant lines al3-4 and al4-7 showed more than 50% decrease in expression (Figure 3C). These mutant lines were subjected to infection essays with virulent as well as avirulent strains of P. syringae DC3000.
Infection essays were performed using virulent strain DC3000 Pto and avirulent strain DC3000 ∆avrPto/∆avrPtoB. In case of DC3000 Pto growth rate was significantly higher in all the tested amiRNA plant lines as compared to wild type (Col-0) plants during all three time points tested (Figure 4A). Whereas in case of DC3000 ∆avrPto/∆avrPtoB growth rate was significantly higher in mutant plants at 1 dpi and there was no significant difference at later time points (Figure 4C). Addition of L-arginine (1 mM) in infiltration medium reduced the growth rate of both P. syringae strains in mutant plants but did not affect the control plants significantly. However, overall growth rate was reduced in both control and mutant lines compared to non-treated plants (Figure 4 B & D).
Figures & Tables
This study was performed to reveal the potential role of relevant genes for important enzymes of arginine metabolic pathway and their relevance in A. thaliana-P. syringae interactions. Presented data clearly indicate the remodeling of gene expression for these genes during P. syringae infection in A. thaliana. Our study shows importance of arginine anabolic genes and enzymes for successful plant defense against P. syringae. Induced gene expression of AS and AL upon P. syringae infection could be the result of defense related strategies employed by A. thaliana. Currently very little information is available about arginine cycle in A. thaliana and only a few studies are centered to this area of research [14]. Even, very less is known regarding its role in plant-pathogen interactions. However, it has been demonstrated that arginine cycling is modulated during compatible plant-nematode interactions [8,15]. Arginine is an important amino acid and centers many metabolic pathways involving plant defense reactions against abiotic [23] and biotic stresses [24,25]. Arginine cycle genes are also found to be modulated during mutualistic relationships of symbiotic fungi with plants [26].
To reveal the potential role of two important genes of arginine pathway, amiRNA-based gene silencing mutants of A. thaliana were generated. Argininosuccinate Synthase was suggested to be rate limiting enzyme of arginine biosynthesis [27]. Based upon these findings, two genes AS and AL were targeted for silencing in arginine metabolic pathway. Silencing of these genes reduced the expression significantly. Subsequently these mutants were subjected to infection assays with P. syringae. Silencing of AS and AL genes increased the growth rate of P. syringae on mutant plant lines. This phenomenon was reversed when L-arginine was added in the infiltration medium. Arginine is found to be essential for the synthesis of nitric oxide (NO) in A. thaliana [12] which is an important molecule as far as plant defense responses are concerned [10]. In our studies decrease in expression of AS and AL results in an increase of the growth of P. syringae on A. thaliana. This change can be traced back to reduced NO production and compromised plant defense. This hypothesis is also supported by the fact that decrease in the AL activity leads to the reduced production of NO [28].
Expression of two arginine catabolic genes associated with arginase enzyme was also studied. Arginase mostly functions to mobilize stored nitrogen from arginine in plants [13]. Arginase expression is also reported to be enhanced by wounding [29], as well as during pathogen infection [30]. Arginase and Nitric Oxide Synthase (NOS) use the same substrate, which is arginine [23]. Increase in the expression of arginine
synthesis genes and not in arginase genes suggests cycling of arginine towards NOS for the synthesis of NO because arginase negative mutants are suggested to accumulate more NO as compared to wild type plants [31].
Our data displays that silencing the two important genes in arginine biosynthesis increases the growth rate of P. syringae in A. thaliana. This could be the result of decrease in arginine production because these two genes are associated with the rate limiting enzymes of arginine cycle. Compromised defense seems to be due to decreased production of NO, which uses arginine as precursor. Addition of arginine in infiltration medium hindered with the growth of P. syringae in mutant plant lines as well as on wild type plants. This finding suggests that sufficient arginine is required for NO-mediated plant defense and its decrease improves the conditions for the growth of pathogen.
Acknowledgement
Higher Education Commission (HEC) of Pakistan is highly acknowledged for providing the funding of this work. Help and kind guidance from Dr. Holger Bohlmann (BOKU, Wien) is also acknowledged.
Authors' Contribution
Nusrat Saba: Designed the study, carried out the genomic work. Saadia Noreen: Performed data analysis. Mubarak Ali Anjum: Participated in sample collection. Saqib Ali: Participated in sample collection. Muhammad Jawad: Participated in sample collection. Allah Rakha: Principle Investigator of the research.
Shahbaz Anwar, Muhammad Amjad Ali, Amjad Abbas and Krzysztof Wieczorek all designed the study, carried out the lab work, drafted the manuscript and corrected the reviewed copy.
- Thordal-Christensen, H. Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology, (2003); 6(4):351-357.
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell, (2006); 124(4):803-814.
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, (2004); 428(6984):764-767.
- Arasimowicz M, Floryszak-Wieczorek J. Nitric oxide as a bioactive signalling molecule in plant stress responses. Plant Science, (2007); 172(5):876-887.
- Ali MA, Abbas A, Kreil DP, Bohlmann H. Overexpression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots. BMC Plant Biology, (2013); 13: 47
- Ali MA, Anjam MS, Nawaz MA, Lam HM, Chung G. Signal Transduction in Plant-Nematode Interactions. International Journal of Molecular Sciences, (2018); 19(6):1648.
- Rai VK. Role of Amino Acids in Plant Responses to Stresses. Biologia Plantarum, (2002); 45(4): 481-487.
- Hofmann J, El Ashry AEN, Anwar S, Erban A, Kopka J, et al. Metabolic profiling reveals local and systemic responses of host plants to nematode parasitism. Plant Journal, (2010); 62(6):1058-1071.
- Anwar S, Wieczorek K, Inselsbacher E. Analysis of Arabidopsis amino acid metabolism in response to Heterodera schachtii infection. Pakistan Journal of Nematology, (2018); 36(2): 131-150.
- Delledonne M, Xia Y, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature, (1998); 394(6693):585-588.
- Imran QM, Hussain A, Mun BG, Lee SU, Asaf S, et al. Transcriptome wide identification and characterization of NO-responsive WRKY transcription factors in Arabidopsis thaliana L. Environmental and Experimental Botany, (2018); 148:128-143.
- Modolo LV, Augusto O, Almeida IMG, Pinto-Maglio CAF, Oliveira HC, et al. Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Science, (2006); 171(1):34-40.
- Micallef BJ, Shelp BJ. Arginine metabolism in developing soybean cotyledons .2. Biosynthesis. Plant Physiology, (1989); 90(2):631-634.
- Slocum RD. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiology and Biochemistry, (2005); 43(8):729-745.
- Anwar S, Inselsbacher E, Grundler FMW, Hofmann J. Arginine metabolism of Arabidopsis thaliana is modulated by Heterodera schachtii infection. Nematology, (2015); 17(9): 1027-1043.
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods, (2011); 7:32.
- Curtis MD, Grossniklaus UA. Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiology, (2003); 133(2):462-469.
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell, (2006); 18:1121-1133.
- Szakasits D, Siddique S, Bohlmann H. An improved pPZP vector for Agrobacterium-mediated plant transformation. Plant Molecular Biology Reporter, (2007); 25(3-4):115-120.
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal, (1998); 16(6):735-743.
- Brownfield D L, Todd C D, Deyholos MK. Analysis of Arabidopsis arginase gene transcription patterns indicates specific biological functions for recently diverged paralogs. Plant Molecular Biology, (2008); 67(4):429-440.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt Method. Methods, (2001); 25(4):402-408.
- Das P, Lahiri A, Lahiri A, Chakravortty D. Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator. Plos Pathogens, (2010); 6(6):e1000899.
- Bouchereau A, Aziz A, Larher F, Martin-Tanguy J. Polyamines and environmental challenges: recent development. Plant Science, (1999); 140:103-125.
- Walters DR. Polyamines and plant disease. Phytochemistry, (2003); 64(1):97-107.
- Anwar S, Wieczorek K. Serendipita indica modulates expression of arginine metabolism genes during colonization of Arabidopsis thaliana roots. Pure and Applied Biology, (2018); http://dx.doi.org/10.19045/bspab.2018.700203
- Haines RJ, Pendleton LC, Eichler DC. Argininosuccinate synthase: at the center of arginine metabolism. International Journal of Biochemistry and Molecular Biology, (2011); 2(1): 8-23.
- Erez A, Nagamani SC, Shchelochkov OA, Premkumar MH, Campeau PM, et al. Requirement of argininosuccinate lyase for systemic nitric oxide production. Nature Medicine, (2011); 17(12): 1619-1626.
- Chen H. Regulation of Plant Arginase by Wounding, Jasmonate, and the Phytotoxin Coronatine. Journal of Biological Chemistry, (2004); 279(44):45998-46007.
- Jubault M, Hamon C, Gravot A, Lariagon C, Delourme R, et al. Differential regulation of root arginine catabolism and polyamine metabolism in clubroot-susceptible and partially resistant Arabidopsis genotypes. Plant Physiology, (2008); 146(4):2008-2019.
- Flores T, Todd CD, Tovar-Mendez A, Dhanoa PK, Correa-Aragunde N, et al. Arginase-negative mutants of Arabidopsis exhibit increased nitric oxide signaling in root development. Plant Physiology, (2008); 147(4):1936-1946.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0