Full Length Research Article
Single nucleotide polymorphism and phylogenetic analysis of the exon 2 of leptin gene in Lohi sheep
Ali Haider Saleem1*, Ahmad Ali2, Akhtar Rasool Asif1, Asad Ali1, Hamid Mustafa1, Dilshad Rashid1, Mujahid Zafar3, Muhammad Awais1, Hafiz Ishfaq Ahmad1, Zulqarnain Baqar1
Adv. life sci., vol. 7, no. 2, pp. 62-65, February 2020
*- Corresponding Author: Ali Haider Saleem (Email: ali.haider@uvas.edu.pk)
Authors' Affiliations
2. National Agricultural Research Center, Islamabad – Pakistan
3. Livestock and Dairy Development Department, Lahore – Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Leptin hormone, encoded by leptin (LEP) gene is involved in many biological and physiological processes in the body. Polymorphism in LEP gene has been observed and correlated with a variety of reproductive and productive traits in several sheep breeds worldwide, but its role has not been much studied in local sheep breeds of Pakistan. The present study was conducted to analyze polymorphism in LEP gene in Lohi breed of sheep.
Methods: Subsequent to statistical analysis (generalized linear model), 18 animals were selected randomly from the flock for blood samples collection followed by DNA extraction, amplification using PCR prior to sequencing. The amplified product of exon 2 and partial intron 2 regions of LEP gene was 268bp.
Results: Molecular analysis showed a heterozygous condition i.e. C>Y at position 15 and 18 in exon 2. The data on average daily weight gain (ADG) from birthday to 90 days were used for association study, while environmental effects were minimized by means of generalized linear model. Association of polymorphisms in LEP gene with ADG did not yield any significant results.
Conclusion: In conclusion, analysis of LEP gene sequence verified the existence of genetic changes in Lohi sheep. Further investigations are needed to find variations that might be linked with traits of economic importance for upcoming breeding program sand marker-assisted selection.
Keywords: DNA; Exon 2; LEP; PCR; Lohi
There are mainly 30 breeds of domesticated sheep inhabiting tropical and temperate areas of Pakistan mostly raised for meat, milk and coarse carpet type wool [1]. Among these, Lohi is an economically superior thin tail sheep breed of province Punjab Pakistan, having large body size [2]. There is an immense variation in development and reproductive traits of this breed which demonstrates a great possibility of enhancement in performance [3]. Numerous hormones are produced in animal’s body and leptin hormone is believed to regulate many important traits related to growth, reproduction and maintenance [4, 5]. It is encoded by LEP gene having three exons and two introns, present on 5th chromosome in sheep [6]. LEP gene is responsible for energy metabolism in the body and it also affects the secretion of gonadotropins and milk in various farm animals [7]. Polymorphisms in the LEP gene and their relationship with growth traits in ovine are reported by many scientists worldwide [8, 9]. Consequently, the aim of the study was to evaluate LEP gene polymorphism and its association with growth trait in Lohi sheep breed, which has not been much carried out in Pakistan.
Sample Collection and Preparation: The data of Lohi sheep were obtained from Small Ruminants Training and Research Centre, Pattoki, Pakistan and statistically analysed to evaluate the effect of environmental factors. The data concerning ADG from birthday to 90 days were obtained and analyzed to conclude the influence of dam’s age, season, sex of new born, type of birth and year of birth. After analysis, 18 animals were randomly selected from the herd, though definite correction factors were also calculated to decrease the effects of environment [10].
Procedure and Analysis: A total of 5 ml blood was collected cautiously from each animal from jugular vein using a sterile syringe and stored in 15 ml centrifuge tubes including ethylenediaminetetraacetic acid (EDTA) as anticoagulant and blood samples were stored at -20ºC. Stored blood samples were subjected to DNA extraction by using a protocol as followed by Babar et al. [11], while primers for LEP gene were designed using Primer3Plus software against NCBI gene accession no: 443534. A primer set, LEP-F “5’-TGGTAACGGAGCTCAT-3’” and LEP-R “5’-TTACCTCATCTCCCTGTC-3’”, was used for the amplification of 268bp fragment covering up the entire exon 2 and partial intron 2 of the gene. Touchdown PCR was performed with a Thermal Cycler (iCyclerBioRad, USA) with initial denaturation at 94ºC for 5 min, followed by 35 cycles of denaturation at 94ºC for 45 sec, annealing 56-46ºC for 45 sec and extension was at 72ºC for 45 sec. Final extension was given at 72ºC for 10 minutes and stored at 4ºC.
Prior to Sanger sequencing from 1st Base Laboratories Singapore, amplicons were cleaned using TIANquick Midi Purification Kit (Tiangen Biotech Beijing Co., Ltd.). The sequences were aligned and edited using CodonCode Aligner ver. 5.0.2 and MEGA 6.0 software [12]. The following model was utilized to minimize the effect of environmental factors:
Yi = µ + Zi + Ɛi
Where, Yi is ADG of nth lamb; µ is overall population mean; Zi is the effect of year, season, sex of new-born, type of birth and age of dam; at the same time Ɛi is a random error associated with ADG of nth lamb. Furthermore, it was presumed that Ɛi was generally and autonomously distributed with mean 0 and variance σ2.
Electrophoretic analysis revealed high molecular weight bands of genomic DNA extracted from blood samples of Lohi sheep on 0.8% agarose gel. The results indicated that extraction method yielded good quality DNA and DNA was suitable for PCR analysis. A PCR was setup for amplification of a specific LEP gene exon 2 segment (268 bp). The PCR created a solitary enhancement from all templates of studied sheep individuals. The extent of all the PCR amplicons demonstrated no undeniable contrasts when isolated on agarose gel (1.8 %) as appeared in Fig. 1. Moreover, amplified DNA fragments were subjected to DNA sequencing to determine the polymorphism in the amplified region.
Figures & Tables
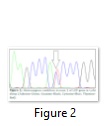
Polymorphism in LEP gene was studied in Lohi animals, an important indigenous sheep breed of Pakistan. Heterozygous peaks were observed in two individuals (ID: 201312025, 201321143) and were labeled as “Y”. The comparative evaluation of individuals carrying “Y” and those lacking it based on phenotypic data did not show any significant association. An attempt was made to correlate observed mutation with ADG but due to a limited number of animals, the analysis did not show any concluding relationship. Qureshi et al. [14] reported variations in intron 2 (A2262T, C2256G, -2730C) and exon 2 (A3201G) in the LEP gene in Lohi sheep maintained at Livestock Production and Research Institute, Bahadurnagar, Okara, Pakistan. Jonas et al. [15] found variation (C11T) in exon 2 in Awassi-Merino sheep with non-significant (P>0.05) association with body weight. Similarly, Boucher et al. [16] also found SNPs in exon 2 of Dorset and Suffolk sheep breeds. Three SNPs were observed by Wang et al. [17] while studying polymorphism in LEP receptor gene in caprine and along-with association between genotypes and traits i.e. birth weights of kids and prolificacy. A relationship was observed between genotypes and weights at different ages of animals in ovines of Makooei breed [18]. Certain SNPs in intron 2 and exon 2 in goats were also reported by Singh et al. [19] and Maitra et al. [20].
Phylogenetic tree (fig. 3) shows that Lohi animals used in this study have conserved regions for exons and goats showed close relation with this. Phylogenetic tree constructed based on exons of leptin gene reconfirmed the biological classification of mammalian species. In other words, molecular classification-based leptin exons gene sequence reconfirmed the classical taxonomical classification. Phylogenetic tree showing inter-relationship of Lohi sheep animals with closely related animal species inferred from exon 2 sequences in ovine LEP gene. Tree was generated using the neighbour- joining methods. Bootstrap values expressed as percentage of 1000 replications and indicated at the nodes.
The present study may be regarded as one of the initial attempts to understand the genetic diversity of LEP gene in local sheep breeds of Pakistan. Molecular analysis of exon 2 of LEP gene in Lohi individuals showed polymorphism. At present, very less data are available to compare our results. There is need to extend this work with a higher number of animals in different breeds to have more candidate polymorphisms that may be associated with growth traits for possible use in Marker Assisted Selection (MAS) in future breeding programs.
Authors' Contribution
AHS conducted the study, AA, ARA and HIA helped in field and lab work, AA and HM did data analysis, DR and ZB helped in drafting, MA and MZ reviewed and approved the draft.
The authors declare that there is no conflict of interest regarding the publication of this paper.
- Ali A, Babar ME, Mahmood S, Imran M. First report of GTG-banded nomenclature of Pakistani Lohi sheep (Ovis aries). Turkish Journal of Veterinary and Animal Sciences, (2011); 35(4): 213-217.
- Fiaz M, Munawar K, Mushtaq M, Ahmad M, Chaudhry MA, et al. Evaluating Varying Calf Milk Replacers for Optimum Growth Performance in Salt Range Lambs. Pakistan J Zool, (2017); 49(5): 1-4.
- Babar M, Ahmad Z, Nadeem A, Yaqoob M. Environmental factors affecting birth weight in Lohi sheep. Pakistan Veterinary Journal, (2004); 24(1): 5-8.
- Singh U, Kumar S, Ganguly I, Gaur G, Jagadeesan K, et al. Identification of genetic polymorphism in two exonic coding region of Leptin gene among indigenous and crossbred cattle. Indian Journal of Animal Research, (2014); 48(5): 403-407.
- Meena A, Bhatt R, Sahoo A, Kumar S. Polymorphism of the exon 3 of leptin gene in Malpura sheep. Indian Journal of Animal Research, (2017); 51(3): 469-473.
- Javanmard A, Mohammadabadi M, Zarrigabayi G, Gharahedaghi A, Nassiry M, et al. Polymorphism within the intron region of the bovine leptin gene in Iranian Sarabi cattle (Iranian Bos taurus). Russian Journal of Genetics, (2008); 44(4): 495-497.
- Priyadarshini L, Yadav AK, Singh HS, Mishra A, Jain AK, et al. Role of leptin in physiology of animal reproduction-A review. Agricultural Reviews, (2015); 36(3): 235-240.
- Bakhtiar R, Abdolmohammadi A, Hajarian H, Nikousefat Z, Kalantar-Neyestanaki D. Identification of g. 170G> A and g. 332G> A mutations in exon 3 of leptin gene (Bcnl and Cail) and their association with semen quality and testicular dimensions in Sanjabi rams. Animal reproduction science, (2017); 179: 49-56.
- Kaplan S, Atalay S. Single Nucleotide Polymorphism of Ovine Leptin and Insulin-Like Growth Factor 1 Gene in Kivircik Crossbred Ewes. Pakistan Journal of Zoology, (2018); 50(3): 851-856.
- Babar ME (1994). GENETICS AND PHENOTYPIC PARAMETERS OF SOME PERFORMANCE CHARACTERISTICS OF LOHI SHEEP: PhD Thesis. University of Agriculture Faisalabad.
- Babar M, Hussain T, Sadia H, Nadeem A, Kamran M, et al. Mitochondrial Cytochrome B gene based phylogeny of Lohi and Thalli sheep breeds of Pakistan. Journal of Animal and Plant Sciences, (2014); 24(4); 1260-1262.
- Saleem AH, Javed K, Babar ME, Hussain T, Ali A, et al. Association of Leptin Gene Polymorphism with Growth Rate in Lohi Sheep. Pakistan Journal of Zoology, (2018); 50(3): 1029-1033.
- Zhang B, Chu L, Liu H, Xie C, Qiao S, et al. Leucine Supplementation in a Chronically Protein-Restricted Diet Enhances Muscle Weight and Postprandial Protein Synthesis of Skeletal Muscle by Promoting the mTOR Pathway in Adult Rats. Engineering, (2017); 3(5): 760-765.
- Iqbal Qureshi Z, Farid AH, Ellahi Babar M, Hussain T. Leptin Gene Polymorphism in Lohi, Kajli and Spili Breeds of Sheep. Pakistan Veterinary Journal, (2015); 35(2):321-324.
- Jonas E, Martin G, Celi P, Li L, Soattin M, et al. Association of polymorphisms in leptin and leptin receptor genes with circulating leptin concentrations, production and efficiency traits in sheep. Small Ruminant Research, (2016); 136: 78-86.
- Boucher D, Palin M, Castonguay F, Gariépy C, Pothier F. Detection of polymorphisms in the ovine leptin (LEP) gene: Association of a single nucleotide polymorphism with muscle growth and meat quality traits. Canadian journal of animal science, (2006); 86(1): 31-35.
- Wang P, Deng L, Zhang B, Chu M, Tan Y, et al. Identification of polymorphism on leptin receptor gene of goats in southwest China. Small ruminant research, (2011); 96(2-3): 120-125.
- Hajihosseinlo A, Hashemi A, Sadeghi S. Association between polymorphism in exon 3 of leptin gene and growth traits in the Makooei sheep of Iran. Livestock Research for Rural Development, (2012); 24(9): 543-546.
- Singh S, Rout P, Agarwal R, Mandal A, Singh S, et al. Characterization of exon 2 and intron 2 of leptin gene in Indian goats. Animal biotechnology, (2009); 20(2): 80-85.
- Maitra A, Sharma R, Pandey A, Singh L, Mandakmale S, et al. Preliminary identification and characterisation of leptin gene polymorphism in Indian goats. Journal of applied animal research, (2014); 42(1): 118-122.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0