Research Article
Molecular detection, phylogenetic analysis and designing of siRNA against Potato Virus X
Shafique Ahmed1*, Idrees Ahmed Nasir1, Tahir Yaqub2, Muhammad Waseem2, Bushra Tabassum1, Faiza Masood2, Anwar Khan1, Shahid Javid Butt3, Tayyab Husnain1
Adv. life sci., vol. 1, no. 1, pp. 37-43, November 2013
*-Corresponding Author: Shafique Ahmed (Email: shafiq.uvas@gmail.com)
Author Affiliations
2-Institute of Biochemistry and Biotechnology, University of Veterinary and Animal Sciences, Lahore – Pakistan
3-Department of Horticulture, PMAS-Arid Agriculture University, Rawalpindi – Pakistan
Abstract
Introduction
Methods
Results
Discussion
References
Abstract
Background: As potato (Solanum tuberosum L.) is one of the most liked food crops for human diet so increasing its production is an important goal for scientists to achieve. In this molecular study, we characterized the Coat Protein (CP) gene of Potato Virus X (PVX). CP gene is virulence mediator and integral part of viral structural assembly.
Methodology: We tissue cultured the PVX positive potato plants for viral RNA extraction. Total RNA was converted to cDNA for priming CP gene in PCR for amplification. To get the complete sequence of gene, we cloned CP gene into pTZ57R/T cloning vector. Upon double digestion of recombinant plasmid with EcoRI and HindIII restriction enzymes, 710 bp fragment was obtained which confirmed cloning. Recombinant plasmid was sequenced with M13 primers.
Results: Derived consensus sequence of 710 bp was found to be exact cds of CP gene showing 95% similarity with referenced genome. Phylogenetic analysis suggested Indian isolate of PVX as the nearest one. Multiple siRNA were designed against mentioned and optimized computationally to provide base for further studies.
Conclusion: Following facts may be established upon findings of this research; i) CP gene of Pakistani isolate of PVX has high homology with other PVX isolates found around the world, ii) in determining target for efficient siRNA mediated approach to silence PVX genome, this conserved nature can be proved very promising. Thus, to develop PVX-resistant potato crop in Pakistan through siRNA mediated strategy, CP gene could be the best target.
Keywords: Solanum tuberosum L., Potato Virus X, Coat protein gene, Sequencing, Phylogenetics, siRNA
Introduction
Potato (Solanum tuberosum L.) comes in fourth position after rice, wheat and maize when its production worldwide is considered. According 2011 report by FAO annual production of potato is with 374.4 million metric. Total area devoted to potato crop is 53,666 thousand hectares around the agriculture world. 159.3 thousand hectares land was utilized for production of 3491.800 thousand tones of potato in Pakistan. (FAOSTAT, 2013). Among major viral pathogen of this crop, there exists Potato Virus X (PVX). This virus is not only common limiter of production in Pakistan but around the globe. Genetically, PVX carry 515 nm long RNA genome as a single positive-strand filamentous stranded. There are five genes complementing full genome namely; 3′-proximal coat protein (CP) gene, triple-gene block’ (TGB) coding for proteins involved in viral moment, and 5′-proximal gene producing 165 k of the Dareplicase [1]. Severe mosaic, dwarfing of the plant, mild mottling of the leaf and reduced leaflet size are the major symptoms shown in potato plants after getting infected with this virus. Solanaceae family of plants serve as its host entities [2]. During a year of average and normal infection, cost of the damage in the United Kingdom to potato crop yield reduction by three virus combined; PVX, Potato Virus Y (PVY) and Potato Leafroll Virus (PLRV) was anticipated between £30 to £50 million [3]. Production yield of potato across Pakistan is generally lowered by 5-10% due PVX [4]. There is no recent study reporting fresh statistics of loss to potato yield caused by PVX. Rapidly mutating genome easily fails the conventional methods of controlling virus [5]. To pave a path toward above stated goal, we must have to survey complete genome of virus and search for the most conserved part against which techniques like small interfering RNA or siRNA could be applied. siRNA has been increasingly used against variety of viral diseases for both fauna and flora [6-8].
Methods
Sampling and tissue culturing of PVX positive potato plants
Based upon typical symptoms of PVX; tuber malformations, mosaic patterns on leaves, stunting of the plant and leaf malformations, PVX positive plants were sampled from potato field. MS basal media no antibiotics or zero media was used to maintain through single node cutting and placed in 16 hrs of photo-periods at 22 ± 2°C. After every two to three weeks, plants were sub-cultured to maintain sufficient supply of positive samples for further experimentation.
Extraction of total RNA and cDNA synthesis
RNA was extracted through 'single-step' method introduced by Chomczyncki (1987) from leaves of the infected potato plants. Transferring of leave cuttings from culture room to lab and grinding of leaves were done in liquid nitrogen to avoid degradation of heat sensitive RNA. For PCR amplification of CP gene, cDNA was synthesized from extracted RNA by “RevertAid first strand cDNA Synthesis kit” (Catt# 1621; Fermentas) according to kit protocol.
PCR optimization and amplification
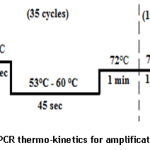
To detect and amplify CP gene of PVX, we designed forward 5’AGTGAGGACTGAACCTTGTGTC3’ and reverse primer 5’ATGAAACTGGGGTAGGCGTC3’using Primer 3 software tool [9]. Primers were tested in-silico (on insilico.ehu.es/PCR) and optimized at 60oC Tm using gradient PCR. Thermo-kinetics during gradient PCR are shown below in figure 1 below.
TA Cloning and Sequencing
TA cloning vector pTZ57R/T (Cat#1213; Fermentas) was used to ligate purified PCR product in 1:1 molar ratio of vector: insert. Overnight (16-20 hrs) incubation at 14oC was given to ligation mixture. Heat shock method was used to transform freshly prepared E. coli DH5α competent cells with ligated product. To screen the positively transformed bacterial colonies, blue/white selection assay was applied. Increase the number of transformed colonies, 5ml of LB broth having ampicillin (50μg/mL) in it, was inoculated with white colonies by the help of sterilized microbiological loop. Overnight incubation was given to inoculated cultures at 37°C. Recombinant plasmids were isolated through “GeneJET Plasmid Miniprep Kit” (Cat#0502; Fermentas) from selected transformed colonies after incubation in broth containing amplicin drug. EcoRI and HindIII enzymes were used to digest the isolated recombinant plasmid DNA for confirmation of the positive clone containing PVX-CP gene. Agarose gel (1%) was used to analyze the digested fragments of recombinant plasmid. After digestion with restriction enzymes, two bands were appeared on gel. Confirmed plasmid was then sequenced with M13 universal primers.
Results
Amplification of the CP gene
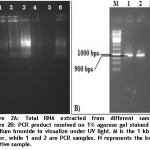
After optimizing primers and thermo-kinetics with the help of gradient PCR, we found that best amplification of the gene was achieved at Tm 60oC. In figures 2A and 2B, RNA extracted from positive leaf samples for further processes and amplified CP gene resolved on 1% agarose gel are shown below.
Cloning and Restriction Analysis
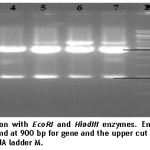
For confirmation of the positive clone containing PVX-CP gene, the isolated recombinant plasmid DNA was digested with restriction enzymes EcoRI and HindIII.
1% agarose gel was used to separate the digested fragments. Digestion resulted in two bands; insert size was 900bp while band of vector was recorded at 2.5kb (figure 3).
Sequencing and Phylogenetic Analysis
Recombinant plasmid was sequenced with M13 universal primers (forward & reverse). Consensus sequence of both primers was generated in Bioedit 7.0.1. Below (in text box 1) is the consensus sequence generated after comparing with sequence of CP gene accessed from reference genome. As CP gene was supposed to be embedded in vector overhangs, so target gene sequence was extracted by checking the homology with reference CP gene available at NCBI. Multiple sequence alignments were done with the nucleotide sequence of PVX Pakistani isolate and obtained significant resemblance with other reported isolates of the same gene. That phylogenetic closeness can be idealized in the phylogenetic tree developed after aligning discovered sequence with other similar sequences.21 sequences meat the selection criteria to get included in phylogenetic tree i.e. query coverage 100% and homology more than 93%. This discovered isolate showed its maximum homology of 95% with Indian isolate (Accession No. JF430080.1) followed by Scottish isolate (Accession No. GU144349.1) having same 95% homology but with less score than Indian isolates clearly shown in the phylogenetic tree (figure 4). It is also evident that CP gene is more conserved as most of the isolates have highly similar sequence having difference of approximately 5%. The result clearly suggested that CP gene is the most efficient candidate, as it is conserved, for molecular detection. Moreover, siRNA designed against this gene could have maximum efficiency. Phylogenetic tree, created by MEGA 4.1 [10] of our local isolate of PVX on the basis of its CP gene is as shown in figure 4.
siRNA Designing
siRNAs were designed against PVX-CP RNA by using 710 bp fragment sequence. To design siRNAs, an online available software tool named as siRNA Wizard v3.1 [http://www.sirnawizard.com/siRNA.php] was used. For designing siRNAs, 50-100 nucleotides after start codon (at downstream region) with 35%-50 % GC contents were selected. Regions sharing sequence homology with other genes of the same organism and having 5 or more nucleotide repeats were not included. All oligos were 21 nt in length. The following 9 siRNA (table 1) oligonucleotides sequences were designed through siRNA wizard tool against CP gene of PVX. 9 predicted siRNA were selected out of total designed oligos on the basis of start position and GC contents percentage. For better efficacy,5' and 3' un-translated regions or UTRs should be avoided.
|
siRNA Sequence |
Start Site |
GC% |
|
GGCTATCTGGAAGGACATGAA |
172 |
47.62 |
|
GCTCAAACAGAAATGATAGAT |
263 |
33.33 |
|
GGCAGCAGCAATCAAAGAGGT |
322 |
52.38 |
|
GGAACTGGATGCTGACTAACA |
387 |
47.62 |
|
GCTGCATTCGACTTCTTCAAT |
461 |
42.86 |
|
GCCACCATCTGAAGCTGAAAT |
529 |
47.62 |
|
GAGGTCGTATCACTGGAACAA |
638 |
47.62 |
|
GGTCGTATCACTGGAACAACA |
640 |
47.62 |
|
GTCGTATCACTGGAACAACAA |
641 |
42.86 |
Table 1: Designed siRNAs against coat protein coding gene of Potato Virus X
Binding sites for regulatory proteins at these regions may hinder the RISC complex accessibility to RNA target sequence [11]. Low GC content (between 30% and 55%) are usually recommended to choose the target sequences [12].These siRNA could be cloned in psiRNA-h7SK G1 expression vector at Bbs1/Bbs1 cloning site.
>PVX_CP_gene_completecds (710 bp) (5’-3’)
ATGTCAGCACAGCTAGCACACAAGCACAGGGTCAACTACTTCACTACACAAAACTGCAGCGCACTCTGGC
ACAGCTCAGACTGTCACATCCTGATGGGGATTTCTTTAGTACAGCCCGTGCTATAGTAGCCAGTAATGCCG
TTGCAACAAATGAGGACTCAGCAAGATTGAGGCTATCTGGAAGGACATGAAGGTGCCCACAGACACTATGGC
ACAGGCTGCTTGGGACTTAGTCAGACACTGCGCTGATGTGGGTTCATCTGCTCAAACAGAAATGATAGATAC
AGGTCCCTATTCCTATGGCACCAGCAGAGCCAGACTGGCAGCAGCAATCAAAGAGGTGTGCACACTCAGGC
AATTTTGCATGAAGTATGCCCCAGTGGTATGGAACTGGATGCTGACTAACAACAGTCCACCTGCTAACTGGC
AAGCACAAGGTTTCAAGCCGGAGCACAAATTCGCTGCATTCGACTTCTTCAATGGAGTCACCAACCCAGCTG
CCATCATGCCCAAAGAGGGACTCATCCGGCCACCATCTGAAGCTGAAATGAATGCTGCCCAAACTGCTGCC
TTTGTAAAGATTACGAAGGCCAGGGCACAATCCAAGGACTTTGCAGCCTAGATGCAGCTGTCACTCGAGGTC
GTATCACTGGAACAACAACCGCCGAGGCTGTTGTCACTCTCCCACCACCATAACTACGTCTACATAA
Text Box 1: Nucleotide sequence of PVX-CP gene
Discussion
In the present study, the first objective was to develop a reliable and reproducible PCR based detection protocol of PVX for commercial purposes. Greater sensitivity of PCR makes it superior technique than molecular hybridization like ELISA. It requires a small sample quantity to diagnose with much faster speed than any other approach. As more plant pathogen are getting their genomes sequenced, so as PCR detection assay against more viriods, bacteria and pathogen is getting developed. The PCR has also been successfully utilized for the detection of plant viruses for example, Grapevine Virus A (GVA) from infected grapevine, apple scar skin and pome fruit virus and Potato Virus A (PVA) in dormant tubers [13]. Simultaneous detection of five potato viruses i.e. potato virus X (PVX), potato virus S (PVS), potato leaf roll virus (PLRV), Potato Spindle Tuber Viroid (PSTVd), PVY and PVA has been done with multiplex RT-PCR (m-RT-PCR) [14]. Real time PCR has been reported with more efficiency than conventional PCR to detect PVX, but it is more costly on account of equipment and reagents involved.
This is why the detection method opted here in this study is conventional PCR. It has also been suggested that the conventional PCR based assay can be considered as a potential and reliable test for routine diagnosis of potato viruses because it showed higher sensitivity as well as time-efficient [15]. So, for characterization studies of PVX in potato crop, we used this molecular technique. Other objective of this study was cloning and sequencing of amplified fragment gene of PVX-CP for characterization of local isolate. Amplified PVX-CP gene was cloned in pTZ57R/T TA cloning vector to achieve the above cited objective. As standard procedure of restriction analysis was performed for confirmation of successful cloning. Recombinant plasmid was sequenced with M13 primers. Homology of derived sequences was tested with other CP genes of PVX reported in NCBI nucleotide database. We found that our sequenced PVX-CP gene showed strong homology (95%) with all reported isolates of PVX. It means that our sequenced gene is relatively conserved with PVX genome that with PVX genome that is similar to previous reports.
Pakistani isolate has very similar sequence of CP gene when compared to Indian isolate and European isolates but somewhat far from Australian isolate in phylogenetic tree. In this study, 710 bp of the total coat protein gene size of PVX was sequenced. Multiple sequence alignments were done with this nucleotide sequence of PVX Pakistani isolate and developed a phylogenetic tree from multiple sequence alignment. Before the studying of multiple sequences alignments and phylogenetic analysis, the homology of the sequenced gene was studied with the already reported sequences in the Gene Bank database through BLAST (Basic Local Alignment Search Tool) software it showed and maximum homology 95% with Indian isolate (Accession No. JF430080.1) and Scottish isolate (Accession No.GU144349.1). On the basis of our findings from sequence analysis, it may be concluded that i) PVX-CP gene has almost same sequence as other PVX isolate do possess and ii) this similarity reflects its conserved nature which in turn could be very helpful to define the siRNA target in PVX genome. These results clearly show that our hypothesis that CP gene could be the best target for siRNA based strategy to develop PVX-resistant potato crop in Pakistan.
References
- Karpova O, Zayakina O, Arkhipenko M, Sheval E, Kiselyova O, et al. Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein–RNA–movement protein’complexes. Journal of general virology, (2006); 87(9): 2731-2740.
- Bostan H, Haliloglu K. Distribution of PLRV, PVS, PVX, and PVY (PVYN, PVYo and PVYc) in the seed potato tubers in Turkey. Pakistan Journal of Biological Sciences, (2004); 7(7): 1140-1143.
- Hull R. Rapid diagnosis of plant virus infections by spot hybridization. Trends in biotechnology, (1984); 2(4): 88-91.
- Rashid F, Khalid S, Ahmad I, Mughal S. Potato virus X (PVX) resistance in tomato cultivars. International Journal of Pest Management, (1989); 35(4): 357-358.
- Lecoq H, Moury B, Desbiez C, Palloix A, Pitrat M. Durable virus resistance in plants through conventional approaches: a challenge. Virus research, (2004); 100(1): 31-39.
- Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. The Plant Cell Online, (2002); 14(4): 857-867.
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. The EMBO Journal, (2002); 21(17): 4671-4679.
- Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic acids research, (2004); 32(19): e149-e149.
- Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Bioinformatics methods and protocols: Springer. pp. 365-386.
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular biology and evolution, (2007); 24(8): 1596-1599.
- Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO reports, (2003); 4(6): 602-608.
- Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS letters, (2002); 532(1): 227-230.
- Hadidi A, Yang X. Detection of pome fruit viroids by enzymatic cDNA amplification. Journal of virological Methods, (1990); 30(3): 261-269.
- Nie X, Singh R. A novel usage of random primers for multiplex RT-PCR detection of virus and viroid in aphids, leaves, and tubers. Journal of virological Methods, (2001); 91(1): 37-49.
- Narayanasamy P (2011) Detection of Virus and Viroid Pathogens in Plants. Microbial Plant Pathogens-Detection and Disease Diagnosis:: Springer. pp. 7-220.