![]()
Molecular diagnostics for foodborne pathogen (Salmonella spp.) from poultry
Awais Afzal*¹, Adeela Hussain¹, Muhammad Irfan¹, Kauser A. Malik¹
Adv. life sci., vol. 2, no. 2, pp. 91-97, February 2015
*- Corresponding Author: Awais Afzal (Email: awaisafzal_90@hotmail.com)
Author Affiliations
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Salmonella species (spp.) are among major food-borne pathogens all over the world. Salmonella typhimurium is the main cause of food poisoning in humans. The fundamental objective of this study is to develop a rapid and reliable method to detect Salmonella (a foodborne pathogen) in raw poultry meat by using molecular approaches.
Methods: Total 200 samples of raw poultry meat were collected from different regions of Lahore and analyzed for the presence of Salmonella spp. fimA gene. Similarly, sent genes were selected for the detection of Salmonella typhimurium and Salmonella enteritidis respectively. PCR technique was optimized for diagnosis of contamination.
Results: Out of 200 samples, 2% samples had shown successful amplification of fimA gene representing the presence of serovar Salmonella typhimurium. PCR assay combined with enrichment can enhance the efficiency for detection of Salmonella in poultry.
Conclusion: A robust, simple and convenient PCR based method has been developed for the detection of one of the major food-borne pathogen Salmonella typhimurium.
Key words: Salmonella typhimurium, Salmonella Enteritidisenteritidis, Raw poultry meat, Contamination, Molecular approach
Introduction
Food-borne pathogens have been implicated in various enteric diseases in human, ranging from sporadic cases to large outbreaks [1, 2]. According to Center for Disease Control and Prevention (CDC), food-borne pathogens cause 76 million infections, 325,000 hospitalizations, and 5000 deaths in USA each year [3]. Infection or food poisoning through poultry meat and eggs pose a great threat for public health due to its high consumption worldwide. Most of the infections caused by poultry meat and egg consumption are caused by Salmonella typhimurium and Salmonella enteritidis. Salmonella subsp. enterica serovar enteritidis and typhimurium are among the most common agents causing diarrhea in domestic and wild animals, humans and rodents [4]. Salmonella has a great ability to multiply in food stuffs where it can survive for several years. S. typhimurium is not only the major cause of diarrhea and food poisoning but it has also been a great risk for cardiovascular, bone and joint infections in humans [5-7].
It has been reported that poultry birds with age less than 3 days were affected by virulent systemic disease due to Salmonella. But, the infection rarely produces clinical manifestations in chicken over 3 weeks of age [8-10] and mortality can be higher in adverse conditions for older chicken [9].
Conventional culture methods for isolation of Salmonella use non-selective pre-enrichment or selective enrichment media followed by plating on different agars. Furthermore, for confirmation of pathogen, biochemical and serological assays are opted [9, 11]. The whole procedure takes more than a half week to complete the diagnostic procedure [10, 12]. Additionally, there are sensitivity and specificity problems including culturing of non-target micro-organisms, detection of false positive and over-looking of pathogen that are present below threshold value. For a rapid, robust and reliable detection with minimum time, certain methods i.e. nucleic acid hybridization, DNA and RNA probing and immune-detection are important for detection of Salmonella [2].
In 1999, the European commission approved Polymerase chain reaction (PCR) diagnostics for foodborne pathogens detection to make the procedure harmonized and standardized [13]. Microbiological diagnostics use PCR as a powerful tool for in vitro amplification of DNA [12]. Multiplex PCR has been validated for simultaneous detection of foodborne pathogens in food samples [14].
The rapid, cost-effective, and automated diagnosis of food-borne pathogens throughout the food chain continues to be a major concern for the industry and public health. The main aim of this study was to validate food safety and develop a rapid and reliable method for detection of Salmonella in poultry. Due to number of restrictions, the conventional approaches for detection of food-borne pathogens are not efficient now. PCR has replaced them largely in developed world. In Pakistan, however, the PCR is not currently used for the detection of food-borne pathogens in poultry, beef and milk samples. The conventional approaches are time consuming, labor intensive and often not reliable in contrast to PCR which is a rapid molecular test with high sensitivity and specificity.
Methods
Collection of Reference Strains
The Salmonella reference strain was obtained from The University of Veterinary and Animal Sciences, Lahore for optimization of growth media and PCR conditions. It was inoculated in tryptic soya broth and Selenite Cystine broth then incubated at 37°C overnight.
Phenotypic Identification of Salmonella spp.
Phenotypic identification of Salmonella spp. was done by streaking pure culture of Salmonella strain on Salmonella Shigilla agar (SS agar) plates followed by overnight incubation at 37°C. It was further confirmed by the biochemical test i.e. Triple Sugar Iron Test (TSI). TSI slant was prepared from triple sugar iron agar with a thick butt in test tubes. A loop full of pure cultured growth (black centered colonies from SS agar) was streaked on the slant gently. The tubes were plugged with cotton plug and incubated at 37°C for overnight.
Isolation of Bacterial Genomic DNA
DNA was isolated by using cetyltriethylmethylammonium bromide (CTAB) method from enriched culture. The quantity and quality of the DNA samples were assessed by resolving on 1% agarose gel by applying 90-110V for 30 minutes [13-15].
Detection of Salmonella Reference Strains by Molecular Methods
For molecular diagnostics, PCR was optimized to detect fimA gene from S. typhimurium and sent gene from S. enteritidis. Primers with unique sequences were designed for targeting both of the genes. The sequence and other data of the primers are given in Table 1. All PCR reagents were supplied by Fermentas USA and all PCR assays were performed in gradient thermocycler (Verities, Applied Biosystems).
Optimization of PCR
PCR reactions were optimized separately for fimA and sent genes. Final volume of PCR reaction mixture was 25 µL containing 1 µL bacterial DNA as template, 2.5 µL of 10X Taq buffer, 2.0 µL MgCl2 (1.5 mM), 0.5 µL of dNTPs (2.5 mM), 1.0 µL of each forward and reverse primer (20pmol), 15 µL of double distilled water and 2.0 µL Taq Polymerase (1.0 U/µL). Thermal cycle conditions were same for both genes as shown in Figure 1. Amplified target DNA fragments were checked on 1.5% agarose gel by applying 90-110V for 45 minutes.
Detection of Salmonella spp. in Poultry Samples
A total of 200 samples of raw poultry chicken were collected from various regions of Lahore. These samples were collected in sterilized falcon tubes containing 15 mL of phosphate buffer saline (PBS) solution and were brought to the research laboratory at Forman Christian University Lahore under cold conditions. The enrichment of the samples was done by inoculating 20 µL of PBS in 10 mL of Selenite Cystine broth followed by incubation at 37°C for 12 hours.
For the detection of virulent Salmonella spp. i.e. typhimurium and enteritidis, the molecular approach (PCR) was implemented. Enriched culture broth and autoclaved distilled water were taken in 1:1 in a PCR tube and boiled at 100°C for 20 minutes. After boiling, 5 µL from this mixture was used as DNA template in each reaction mixture of 25 µL. Identification of fimA gene of serovar typhimurium and sent gene of serovar enteritidis were done by using specific primers. PCR amplified products were analyzed on 1.5% agarose gel by applying 90-110V for 45 minutes. Thermal cycler conditions were used as described earlier in Figure 1.
Results
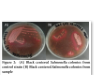
Pure culture of reference strain gave typical black centered colonies on SS agar (Figure 2a). While, among 200 samples of raw poultry meat only 1 (0.5%) gave Salmonella colonies (Figure 2b).
These colonies were then incorporated for further confirmation of Salmonella spp. using biochemical tests. TSI was performed which gave red colour at the surface of slant and black colour in the butt. Salmonella spp. ferments the glucose but do not ferment the sucrose or lactose. Aerobic surface of the slant becomes red due to the increase in pH as they use the protein as source of carbon. Black colour is produced due to hydrogen sulfide. The result is shown in Figure 3.
PCR was used to detect Salmonella spp. from poultry meat samples. DNA of the positive control strain was isolated by using CTAB method. DNA was dissolved in autoclaved distilled water and was checked on 1% garose gel. PCR was performed using the DNA of positive control strain as template.
The PCR products were observed on 1.5% agarose gel. 50bp ladder was used to estimate the product size. A positive PCR showed the amplified products of both of the genes fimA (314bp) of S. typhimurium and sent (310bp) gene of S. enteritidis (Figure 4).
Total 200 samples of raw poultry meat were used for rapid detection method by using molecular approaches. The enrichment of samples were done for overnight and the presence of Salmonella enterica serovar typhimurium and serovar enteritidis was checked directly from enriched samples by applying PCR for their specific genes fimA and sent respectively. The PCR products were observed by electrophoresis on 1.5% agarose gel. Out of 200, 4 (2%) samples represented the presence of serovar typhimurium. The results are shown in Figure 5 and Table 2.
Tables & Figures
Figure 1 Figure 2 Figure 3 Figure 4 Figure 5 Table 1 Table 2





Discussion
S. enteritidis and S. typhimurium are the most common serotypes from Salmonella spp.that are widely considered as disease causing agents in humans [15, 16]. Poultry has always been the most important reservoir which causes the transmittance of Salmonellae to humans. The detection of Salmonella spp. takes nearly a week to confirm the presence of pathogens using conventional microbiological techniques [17]. It is essential to identify and detect the pathogens in short period of time during outbreaks for the public health and safety [18]. The main aim of the present study was to develop a sensitive, robust and simple PCR based method to identify and detect S. typhimurium and S. enteritidis in raw poultry meat and chicken products from local sources.
Conventionally, detection of Salmonella is done by using media such as SS agar, which are largely considered as time-consuming and require additional testing which is very expensive [19]. Rapid detection is achieved by using PCR technique. It is found that PCR is far better in specificity and sensitivity than conventional methods, as conventional methods for Salmonella spp. have reported numerous false positive results [20, 21]. Previous studies successfully detected Salmonella in 11.8% of the samples using PCR based assay [22], whereas traditional culture methods detected only 2.6% of the pathogen from the same samples [11].
Over the last decade, it has been reported that enrichment of various food samples provided more efficient results when tested by PCR rather than previously used culture techniques [22]. It has been reported that more sensitive genus level detection of Salmonella is done by PCR test combined with selective enrichment. Enrichment method increased the number of required viable count as dead organisms reduces the probability of detection and even 2 μL of enrichment was able to produce the PCR results which indicate the PCR sensitivity and specificity [23].
Previous studies reported that when DNA was extracted without pre enrichment, it was difficult to detect Salmonella in food due to its presence in low quantity [21, 24]. In this study, when DNA of Salmonella was directly isolated without enrichment it did not give amplification product. Therefore, bacterial DNA was extracted from poultry samples after enrichment and then amplified. Previous studies [24, 25] reported that when DNA is extracted without pre enrichment step, it is really difficult to detect Salmonella in food due to its presence in low numbers. For that matter, we performed enrichment of samples prior to DNA extraction. DNA extraction protocol was followed reported in various literatures [26-28]. Selenite cysteine selective enrichment is used to increase the specificity and sensitivity of PCR to detect Salmonella spp. in the collected samples.
In this study, by using conventional detection methods of culturing and biochemical tests, (1/200) 0.5% of Salmonella spp. was found. Traditional detection of Salmonella is done by culturing on selective media. The characterization of suspicious colonies is further done by biochemical tests [22].
In present study, S. typhimurium was detected in 2% of the samples taken from the poultry indicating the presence of the pathogens in poultry of Lahore region. In this study, 4 isolates were detected as Salmonella isolates (by using PCR detecting fimA gene). In PCR reaction, fimA gene was amplified and used as a positive test for S. typhimurium. It was selected on the basis of results obtained from preceding study [23]. The fimA gene consists of unique sequences which are reported only in S. typhimurium isolates and so suggests that for the successful detection of S. typhimurium via PCR, this gene is the most suitable target [29-32]. The PCR assay for the identification of S. typhimurium amplified a 314bp fragment of fimA gene which is visualized by gel electrophoresis while DNA from S. enteritidis did not produce any amplified product. In this study, no isolates belonging to S. enteritidis (by using PCR detecting sent gene) were found. sent gene is one of the most suitable targets for the identification of S. enteritidis isolates [32].
PCR procedure has enabled us to not only recognize the organism but also be used as a screening tool as it also helps to indicate the presence or absence of the pathogen.
In conclusion, it is proved that the PCR assay is one of the most highly specific, sensitive and time saving technique for the detection of Salmonella from poultry. PCR assay combined with enrichment step can enhance its efficiency and detection of more accurate presence of Salmonella in poultry. This method is helpful to obtain accurate results in hours rather than weeks as in standard microbiological techniques and biochemical tests. Application of this method will be very useful in food industries and large benefits can be attained by this optimized method.
Acknowledgements
Endowment Fund Secretariat, University of Agriculture, Faisalabad
References
- Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C. An overview of foodborne pathogen detection in the perspective of biosensors. Biotechnology Advances, (2010); 28(2): 232-254.
- Zhao C, Ge B, De Villena J, Sudler R, Yeh E, et al. Prevalence of Campylobacter spp. Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from Washington. Applied Environmental Microbiology, (2001); 67(12): 5431-5436.
- Center of Disease Control. Surveillance for Foodborne-Disease Outbreaks, United States. Surveillance Summaries, MMWR. (2006); 55: 01-10.
- Mead S, Slutsker L, Dietz V, Mc-Caig L, Bresee J, et al. Food-related illness and death in the United States. Emerging Infectious Diseases, (1999); 5(5): 607-625.
- Beal P, Wigley C, Powers B, Hulme D, Smith A. Age at primary infection with Salmonella enterica serovar Typhimurium in the chicken influences persistence of infection and subsequent immunity to re-challenge. Veterinary Immunology and Immunopathology, (2004); 100(3): 151-164.
- Bohez L, Ducatelle R, Pasmans F, Bottledorn N, Haesebrouck F, et al. Salmonella enterica serovae Enteritidis colonization of chicken caecum require the HilA regulatory protein. Veterinary Microbiology, (2006); 116 (1): 202-210.
- Lan R, Stevenson G, Donohoe K., Ward L, Reeves P. Molecular markers with potential to replace phage typing for Salmonella enterica serovar typhimurium. Journal of Microbiological Methods, (2007); 68 (1): 145-156.
- Ocepek M, Pate M, Micunovic J, Bole-Hribovšek V. Comparison and optimization of two PCR tests for identification of Salmonella in poultry feedstuffs, liver and feces. Slovenian Veterinary Research, (2006); 43 (1): 61-66.
- Calnek W, Barnes H, Beard C, Reid W, Yoder H Jr. Diseases of poultry, (1991); 9: 99-120. Iowa State University Press, Ames, Iowa, USA
- Beal P, Wigley C, Powers B, Smith A. Cross reactive cellular and humoral immune responses to Salmonella enterica serovar Typhimurium and Enteritidis are associated with protection to heterologous re-challenge. Veterinary Immunology and Immunopathology. (2006); 114(1): 84-93.
- Van Kessel S, Karns J, Perdue M. Using a portable real-time PCR assay to detect Salmonella in raw milk. Journal of Food Protection, (2003); 66(10): 1762-1767.
- Malorny B, Hoorfar J, Hugas M, Heuvelink A, Fach P, et al. Interlaboratory diagnostic accuracy of a Salmonella specific PCR-based method. International Journal of Food Microbiology, (2003b); 89(2): 241-249.
- Malorny B, Tassios PT, Rådström P, Cook N, Wagner M et al. Standardization of diagnostic PCR for the detection of foodborne pathogens. International Journal of Food Microbiology, (2003); 83(1): 39-48.
- Li Y, Zhuang S, Mustapha A. Application of a multiplex PCR for the simultaneous detection of Escherichia coli O157:H7, Salmonella and Shigella in raw and ready-to-eat meat products. Meat Sciences, (2005) 71(2): 402–406.
- McDonough J, Sanders W, Porter G, Kirchman L. Depth distribution of bacterial production in a stratified lake with an anoxic hypohmnion. Applied Environmental Microbiology, (1986); 52 (5): 992.
- Aktas Z, Martin D, Kayacan B, Diren S, Threlfall J. Molecular characterization of Salmonella Typhimurium and Salmonella Enteritidis by plasmid analysis and pulse-field gel electrophoresis. International Journal of Antimicrobial Agents, (2007); 30(6): 541-545
- Bhagwat A and Lauer W. Food borne outbreaks in raw produce can be prevented. Food Quality. (2004); 11: 62–63.
- Stone G, Oberst D, Hays P, McVey S, Chengappa M. Detection of Salmonella serovars from clinical samples by enrichment broth cultivation-PCR procedure. Journal of Clinical Microbiology, (1994); 32(7): 1742–1749.
- Dijk S, Marjan j, Gijs J, Ruijis M. Evaluation and implementation of a chromogenic agar medium for Salmonella detection in stool in routine laboratory diagnostics. Journal of Clinical Microbiology, (2009); 47(2): 456-8
- Dusch H, Altwegg M. Evaluation of new plating media for isolation of Salmonella species. Journal of Clinical Microbiology, (1995); 33(4): 802-804.
- Perez C, Rivera S, Pirela A, Rincon H, Mavarez Y, Roman R. Isolation of Salmonella in poultry carcasses and evaluation of the effectiveness of different enrichment and selective media. Revista Cientifica-Facultad De Ciencias Veterinarias, (2004); 14(2): 177-185.
- Karns S, Van Kessel S, McCluskey J, Perdue L. Prevalence of Salmonella enterica in bulk tank milk from US dairies as determined by polymerase chain reaction. Journal of Dairy Science, (2005); 88(10): 3475-3479.
- Oliveira D, Rodenbusch C, Cé M, Rocha S, Canal C. Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Letters in Applied Micriobiology, (2003); 36(4): 217-221.
- Waltman D. Methods for the cultural isolation of Salmonella. In: Wray C, Wray A. (eds.). Salmonella in Domestic Animals. (2007): 355-372. CABI Publishing, New York.
- Liming H, Bhagwat A. Application of a molecular beacon-real-time PCR technology to detect Salmonella species contaminating fruits and vegetables. International Journal of Food Microbiology, (2004); 95(2): 177-187.
- Meng J, Zhao S, Doyle P, Mitchel E, Kresovish S. Polymerase chain reaction for detecting Escherichia coli O157:H7. International Journal of Food Microbiology, (1996); 32(1): 103-113.
- Lampel A, Orlandi A, Keonegay L. Improved template preparation for PCR-based assays for detection of food-borne bacterial pathogens. Applied Environmental Microbiology, (2000); 66(10): 4539-4542.
- Perelle S, Dilasser F, Malorny B, Grout J, Hoorfar J, et al. Comparison of PCR-ELISA and LightCycler real-time PCR assays for detecting Salmonella spp. in milk and meat samples. Molecular and Cellular Probes, (2004); 18(6): 409-420.
- Alvarez J, Sota M, Vivanco B, Perales I, Cisterna R, et al. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. Journal of Clinical Microbiology, (2004); 42(4): 1734–1738.
- Jawad K, Al-Hamadani H. Detection of fimA and fimC genes of Salmonella isolates by using Polymerase Chain Reaction. Journal of Basrah Researches, (2011); 37(4): 27-36.
- Cohen H, Mechanda M, Lin W. PCR Amplification of the fimA Gene Sequence of Salmonella typhimurium, a Specific Method for Detection of Salmonella spp. Applied and Environmental Microbiology, (1996); 62(12): 4303-4308.
- Drahovska H, Turoa J, Piknova Y, Kuchta T, Zitasova I, et al. Detection of Salmonella by polymerase chain reaction targeted fimC gene. Biologia Bratislava, (2001); 57 (6): 611 – 616.




