Potent Implications of miRNA in Cancer Biology – A Brief Review
Zeeshan Javed, Muhammad Zaheer Iqbal, Muhammad Umair Latif*, Hafiz Muhammad Farooq Yaqub, Qamar Raza Qadri
Adv. life sci., vol. 2, no. 3, pp. 106-109, May 2015
*-Corresponding Author: Muhammad Umair Latif (Email: m.umairlatif3@gmail.com)
Author Affiliations[Date Received: 18/02/2015; Date Revised: 05/05/2015; Date Published Online: 25/05/2015]
Abstract![]()
Introduction
Methods
Discussion
Conclusion
References
Abstract
Groundbreaking findings through high-throughput technologies have deepened our understanding on an intricate interplay between products of coding sequences and noncoding RNAs. Increasingly it is being realized that miRNA, produced from what was previously considered as "genomic trash" have revolutionized the field of molecular and translational oncology. Overwhelmingly accumulating in-vitro and in-vivo studies have demonstrated that miRNA are key players involved in post-transcriptional regulation of gene network in numerous human pathologies including cancer. In this review we have attempted to provide recent advancements related to multifaceted roles played by miRNA in modulation of oncogenes and tumor suppressor genes.
Key words: Cancer, miRNA, Biogenesis, Tumor suppression
Introduction
Cancer is a multistep disease in which genetic or epigenetic alterations produce such cells that significantly perturb growth cycles. As the cancerous cells grow abnormally in number and size, they also become resistant to cell senescence and apoptosis [1, 2]. Furthermore, if these abnormal cells start invading and colonizing neighboring tissues, they may also lead to organ failure [3, 4]. All the activities from differentiation to death, being performed in a cell (normal/cancerous) are strictly based on the central dogma. The process of central dogma is strictly guided and controlled by non-coding RNAs (ncRNA) such as small interference RNA (siRNA) or microRNA (miRNA). Slight deregulations in the levels of these different types of RNAs may lead to malignancy but knowledge about these regulators may be helpful to invent new therapies as well. In the next sections, authors will focus on the role and involvement of miRNA in the formation of cancerous cells.
miRNA have been first reported in year 2002, and since then extensive research has been carried out on these cellular entities to get an insight of their role in various cellular processes [5]. They have been recognized among some key regulators of differentiation [6], proliferation, metastasis and apoptosis of cancer cells [7, 8]. miRNA has also a key role in regulating gene expression at post-transcriptional level and establishing cell’s identity [9, 10]. Therefore, impaired miRNA functioning results in aberrant gene expression that promotes abnormal cell growth and differentiation thus can serve as a hallmark of cancer [11].
Methods
Literature search strategy and selection criteria
In this study, the level of data submission and MIAME compliance was reviewed for 32 articles that included miRNA associations with cancer profiling. Articles published from 2000 to 2015 were picked from well-known journals—PLOS ONE, the Journal of Biological Chemistry, Blood, Science, Clinical Chemistry, Nature and Cell.
Discussion
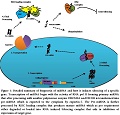
miRNA Biogenesis
miRNA is a ncRNA that is transcribed by RNA polymerase II enzyme. It is synthesized as a single poly-cistronic unit [12] and consists of 19-25 nucleotides [13]. A 2/3rd of all miRNA share a common locus, while
the rest are transcribed as single miRNA [14]. Initially, it is produced as a primary miRNA consisting of 5’ cap structure and a poly A tail [15]. Primary miRNAs are further processed by RNase III enzyme DROSHA and DGCR8 that splice the primary miRNA to pre-miRNA [16]. The pre- miRNA which are double stranded are then exported out of the nucleus by nuclear exporting proteins called exportin-5 [17]. The energy required for this process comes from RAN-GTPase that facilitates this transportation [18]. In cytoplasm pre-miRNA forms complex with DICER and Arguanate (Ago) proteins which then attracts RNA induced silencing complex (RISC) loading complex (RLC). RISC in RLC then cleaves and transforms pre- miRNA into mature miRNA [19]. The mature miRNA is then loaded on to the RNA induced splicing complex which binds to the 3’ UTR region of targeted gene rendering it’s efficient expression [20].
Not all expressed genes are under the regulation of miRNA. Only 30% of whole transcribed genome is being regulated by miRNA [21]. The complex behavior of miRNA gene silencing can be addressed by using various Bioinformatics approaches. These tools have shown that seed region or sequence plays important role in the process of gene silencing by miRNA. Seed region lies at 5’ end of miRNA and requires 100% complementarity with a certain region of mRNA. In case of low match percentage of seed region with specific region of mRNA the process of gene suppression or silencing becomes inefficient. Once this process loses control there are altered levels of miRNA expression, suggesting disturbed cell homeostasis or even cancer [22,23].
miRNA and Cancer
Pertaining to cancers miRNA can be divided into two categories. One that promotes tumor growth and prevent apoptosis from occurring are termed as oncomiRs. While, miRNA that promotes apoptosis and inhibit tumor cell viability, metastasis and invasive abilities are termed as tumor suppressor miRNA [24]. Along with these two categories miRNA manufacturing machinery has been implicated to play a vital role in cancer. The role of miRNA as tumor suppressor was first identified in 2002 when Bulrich et al identified leukemia associated gene (LEU) that contained coding sequences for miRNA in its first intron [25]. Cummino et al using reporter assay in chronic lymphocytic leukemia patients showed that low level of miRNA-15a and miRNA-16 were involved in down regulation of BCL2 gene expression. Their findings have shown that these two miRNA are involved in inversely down-regulating the expression of BCL2 and inducing apoptosis in chronic lymphocytic leukemia derived cell lines [26].
Let-7 is another class of miRNA family which has been implicated to promote tumor suppression in variety of neoplastic as well as solid tumors. Their roles have been extensively studied in lung cancer. High level of Let-7 expression was found to be mandatory for a larger life span in patients suffering from lung cancer [27]. Investigators have revealed the fact that Let-7 directly targets RAS family of proteins inhibiting their expression rendering apoptosis. Mutation in RAS protein effects downstream signaling that results in activation of K-RAS and proto-oncogenic signaling. Let-7 binds to 3’UTR region of K-RAS reducing its efficiency to initiate oncogenic activity [28]. miRNA-34 is another clan of tumor suppressor miRNA that have a well-established role in inhibition of tumor progression. Welch et al first brought the role of miRNA-34 in to consideration by establishing their anti-tumor role in neuroblastoma [29]. p53 is a transcription factor that is active in case of DNA damage and its activity has a direct control over miRNA-34 gene expression. p53 gene binds miRNA-34 in its promoter region thus p53 directly up regulates the expression of miRNA [30].
The anti-tumor activity of miRNA has established them as a useful tool against cancer [31]. Owing to their rich involvement in maintaining cell’s identity they can be constructed as a massive weapon against cancer. The deep insight into the miRNA biology promises great benefits for both diagnosis as well as prognosis of cancer. With the advances in the molecular techniques like nanotechnology, bioinformatics and micro-array the picture depicting miRNA as a sole tool against cancer is becoming clearer and brighter. The therapy relating miRNA as cancer preventive drug are under clinical trials with their own pros and cons nevertheless miRNA mimics based developed drugs are a great hope against cancer.
Post-transcriptional regulation of genes by miRNA has emerged as one amongst the most deeply studied biological mechanism. Data obtained through high-throughput sequencing technologies has considerably enhanced our understanding of tumor suppressing activities of different miRNA in xenografted mice [32].
Conclusion
Rapidly accumulating experimentally verified pre-clinical studies are recommending clinical testing of tumor suppressor miRNA. In line with this approach, miRNA-34 has entered into clinical trials and future studies must converge on developing a broader landscape of signaling landscape of different cancers. Natural and synthetic agents mediated modulation of different miRNA is also an essential dimension and needs further research. Bioavailability and protection of in-vivo delivered miRNA is also a stumbling block that needs to be overcome and recent breakthroughs in gene delivery methodologies will prove to be effective in improving the bioavailability with lesser side effects.
References
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, (1993); 75(5): 855-862.
- Marchetti C, Gasparri ML, Ruscito I, Palaia I, Perniola G, et al. Advances in anti-angiogenic agents for ovarian cancer treatment: The role of trebananib (AMG 386). Critical Reviews in Oncology/Hematology, (2015); 94(3): 302-310.
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell, (2000); 100(1): 57-70.
- Farooqi AA, Wang Z, Hasnain S, Attar R, Aslam A, et al. Citrus Fruits and their Bioactive Ingredients: Leading Four Horsemen from Front. Asian Pacific Journal of Cancer Prevention, (2015); 16(6): 2575-80.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, (1993); 75(5): 843-854.
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Reviews Molecular Cell Biology, (2008); 9(3): 219-230.
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nature Structural & Molecular biology, (2008); 15(3): 259-267.
- Qureshi MZ, Romero MA, Attar R, Javed Z, Farooqi AA. TRAIL and Bortezomib: Killing Cancer with Two Stones. Asian Pacific Journal of Cancer Prevention, (2015); 16(4): 1671-1674.
- Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nature structural & molecular biology, (2008); 15(3): 268-279.
- Farooqi AA, Sarkar FH. Overview on the complexity of androgen receptor-targeted therapy for prostate cancer. Cancer Cell International, (2015); 15(1): 7.
- Dimopoulos K, Gimsing P, Grønbæk K. Aberrant microRNA expression in multiple myeloma. European Journal of Haematology, (2013); 91(2): 95-105.
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, (2001); 294(5543): 858-862.
- Ambros V. The functions of animal microRNAs. Nature, (2004); 431(7006): 350-355.
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Research, (2004); 14(10a): 1902-1910.
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science, (2010); 328(5986): 1694-1698.
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, et al. ATP-dependent human RISC assembly pathways. Nature Structural & Molecular Biology, (2010); 17(1): 17-23.
- Bohnsack MT, Czaplinski K, Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA, (2004); 10(2): 185-191.
- Xie M, Li M, Vilborg A, Lee N, Shu M-D, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell, (2013); 155(7): 1568-1580.
- Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA, (2005); 11(4): 459-469.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature, (2005); 435(7043): 834-838.
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17∼ 92 family of miRNA clusters. Cell, (2008); 132(5): 875-886.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, (2005); 120(1): 15-20.
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell, (2006); 9(3): 189-198.
- Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, et al. Characterization of the 13q14 Tumor Suppressor Locus in CLL Identification of ALT1, an Alternative Splice Variant of the LEU2 Gene. Cancer research, (2001); 61(18): 6640-6648.
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America, (2005); 102(39): 13944-13949.
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, et al. let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell, (2007); 131(6): 1109-1123.
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. RAS is regulated by the let-7 microRNA family. Cell, (2005); 120(5): 635-647.
- Welch C, Chen Y, Stallings R. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene, (2007); 26(34): 5017-5022.
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Current Biology, (2007); 17(15): 1298-1307.
- Lin X, Farooqi AA, Ismail M. Recent progress in fungus-derived bioactive agents for targeting of signaling machinery in cancer cells. Drug Design, Development and Therapy, (2015); 91797.
- Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, et al. Identification of metastasis‐related microRNAs in hepatocellular carcinoma. Hepatology, (2008); 47(3): 897-907.
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Molecular Therapy, (2011); 19(6): 1116-1122.