![]()
Molecular confirmation of Bdv2 gene in wheat germplasm and its field based assessment for resistance against barely yellow dwarf viruses
Samra Kausar1, Shahid Hameed2, Irfan ul Haque3, Kamran Saleem*4, Madiha Zamurrad3, Muhammad Ashfaq3
Adv. life sci., vol. 3, no. 1, pp. 16-22, November 2015
*- Corresponding Author: Kamran Saleem (Email: Kammmran@hotmail.com)
Authors' Affiliations
2- CDRI, NARC Islamabad – Pakistan
3- PMAS-Arid Agriculture University Rawalpindi – Pakistan
4- Nuclear Institute for Agriculture and Biology (NIAB) Faisalabad – Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
Supplementary Data
References
Abstract
Background: Barley yellow dwarf in wheat is an important viral disease among wheat cultivating areas of the world. It is gradually progressing as a major threat to wheat crop in Pakistan due to availability of favorable environmental conditions. The use of resistant cultivar is environmentally safest method for disease control so, it is necessary to develop resistant cultivars before epidemic outbreaks.
Methods: The most commonly used wheat variety Inqilab91 was crossed with BYDV / CYDV resistant variety TC14. F1 generation obtained from the P1 cross was then allowed to self-cross. 61 plants were selected from F2 generation on the basis of disease tolerance or susceptibility and only tolerant plants were included for further experiments in the study. The presence of BDV2 among F2 generation was confirmed by sequence characterized amplified region (SCAR) markers in 6 (91, 96, 110, 119, 121 and 131) out of 61 genotypes which were then backcrossed with recurrent parent. Advance analysis regarding the presence of resistance source among the selected F2 generation was carried out using ELISA. Moreover, appearance of symptoms, agronomic values for different parameters, green house and field responses were also kept under consideration to characterize and confirm the presence of BDV2 among plants.
Results: Results indicated that majority of F2 segregating population showed less yellowing, low viral titer and good agronomic values. ELISA value, glasshouse and field analysis showed that seven genotypes (30, 81, 89, 91,101,110 and 121) were resistant, and twenty-four genotypes were found moderately resistant. Tolerance was detected in genotypes 31, 47, 48, 50, 52, 56, 57, 60, 61, 94, 103, 106, 113, 115, 127, 140 and 413.
Conclusion: Wheat lines containing Bdv2 genes showed resistance in both field and glasshouse. These wheat germplasm could be used as a source of resistance in CDRP-NARC for the further development of resistant wheat varieties against BYDV / CYDV.
Keywords: Virus, Epidemic, resistant gene, ELISA, Marker
Introduction
Wheat provides 21% of the food calories and 20% of the protein to more than 4.5 billion people in 94 developing countries [1]. Therefore, it is ranked fifth regarding usage as staple food, second for major calorie source and first as a protein source Wheat is a leading food grain of Pakistan and being staple diet of inhabitants, it occupies a central position in the agriculture system [2]. Among viral diseases, barley yellow dwarf disease is the most important viral disease of wheat reported in 1953 [3]. It has a worldwide distribution and infects a wide range of Poaceae family including wheat, barley, oats and occasionally rice and maize [4]. In Pakistan, barley yellow dwarf virus (BYDV) was detected in 1992 [5]. Barley yellow dwarf disease is caused by several species from two genera in the family Luteoviridae: Luteovirus (Barley yellow dwarf virus [BYDV-MAV and BYDV-PAV]) and Polerovirus (Cereal yellow dwarf virus [CYDV-RPV, formerly BYDV-RPV]) and three unassigned species (BYDV-RMV, BYDV-SGV, and BYDV-GPV) [6]. They are serologically distinguishable [7]. BYD viruses are not mechanically transmissible, nor through seed but transmitted by aphids in a persistent, circulative but non-propagative manner. Common effects of the virus on agronomic characteristics include reduction in yield, yield components, height, above ground dry weight and root growth [8-11].
Control of the disease can be partially achieved through the application of insecticides, cultural practices (such as changes in sowing date, alternate cropping, and removal of virus reservoirs), and the use of germplasm with tolerance or resistance to the virus or its vectors. The use of cereal cultivars, resistant to BYDV is an effective and cheap means of minimizing the losses caused by BYDV [12]. Generally, high levels of BYDV resistance have been found in barley. The main source of BYDV resistance used in barley breeding programs has been the Yd2 gene, originating from a number of Ethiopian barleys [13]. The BDV2 gene is responsible for imparting resistance against disease in wheat and it’s another wild relative Thinopyrum intermedium. The difference lies between the location and mode of action of the gene in two plants. In wheat this gene is present on homeologous chromosome group 7 (7Ail) while for other it is located on group 2 chromosomes (2Ai-2) [14-19]. The mechanism of BYDV resistance conferred by T. intermediumis is the interference with virus multiplication [20] or reduced cell-to-cell movement [21].
Several molecular markers to detect these translocations are available and have been used for marker-assisted selection [22, 23]. Main cultivars released with Bdv2 include the winter wheat Mackellar (with TC14) and the spring wheat cultivar Glover (with TC6). Currently, none of the Pakistan’s commercial wheat varieties possess resistance against BYDV both under natural and controlled conditions. There is a need to have true resistance against BYDV/CYDV for which identification of Bdv2 gene in ELITE commercial wheat varieties is necessary. Therefore, the present study was initiated to track Bdv2 genes in advance wheat lines through PCR amplification of respective molecular markers and evaluating germplasm containing Bdv2 gene for resistance under controlled inoculation conditions.
Methods
Wheat Germplasm
For introgression of Bdv2 gene, TC14 plant was chosen as a donor parent because it carries the Bdv2 on chromosome 7D [24] and Inqilab91 was chosen as a recurrent parent. Inqilab91 has many genes for adaptation, and gives high yield but it is susceptible for BYDV. During 2007, ten populations of F1 hybrids were developed in field by crossing TC14 x Inqilab91. The F1 hybrids were allowed to self to produce F2 population in glass house. 61 F2 plants were selected at random for field evaluation and confirmation of Bdv2 gene in them by using sequence characterized amplified region (SCAR) marker.
Virus Isolates and BYDV Inoculation
The virus isolates used in these experiments were BYDV-PAV, BYDV-MAV, and CYDV-RPV. The presence of these viruses in field as well as inoculated wheat plants were confirmed through ELISA and PCR in the same laboratory [24-26]. Artificial inoculation was done by depositing 10 aphids on each one-week old infected oat seedling for a 48 hrs transmission period. Aphids acquired the virus on BYDV /CYDV infected oat for a 48 hrs acquisition period. In glasshouse, all wheat lines and oat were sown in 13 cm pots in growth room at temperature 22°C day and 18°C night with a 14 hrs photoperiod. Viruliferous aphids were placed on oat and wheat F2 plants to produce infection. Symptoms on oat and F2 plants was observed after 21 days of inoculations.
Resistance Evaluation in Field Conditions
All 61 F2 wheat plants, evaluated in glasshouse, were also assessed for BYDV resistance in field conditions. Field trials were conducted during 2008-2009 in the research area of CDRP- NARC. The plants were sown in one meter long row having 10 plants in each row along with TC 14 plants as negative control (resistant) and Inqilab 91 (susceptible) as a positive control alternating with susceptible oat rows. Randomized Complete Block Design (RCBD) with three replications was used and whole experimental area was isolated by border rows of oats. All plants were scored according to 0-9 disease rating scale [27] after 45 days of inoculation. Moreover, other parameters such as, number of tillers per plant, plant height, grain weight and finally double sandwiched ELISA was used to determine the success of infection of BYDV.
Primers Designing for Bdv2 Gene and DNA Extraction
Two sets of primers were used to assess the presence or absence of the Bdv2 in segregating lines of F2. One set of primers (BYAGi Forward: 5'- CAT GGA TAA TTC AGG GAG CAT TCT G -3' and BYAGi Reverse: 5' – CTG AAC ACG AAT TTG CTG AGG TTG -3') reported in literature [23] for diagnosis of Bdv2 gene was used. One new set of primer (BDF5Forward: 5′- TGG CAG GCT CGT CGT AC -3′ and BDR6 Reverse: 5′- CTG AGG TTG AGG TTG AGG C -3′) was also designed from Bdv2 gene sequences available in NCBI database. For DNA extraction of wheat leafs, alkali denaturation method was used [28].
Detection of Bdv2 Gene through Polymerase Chain Reaction (PCR)
The reaction mixture was contained in a total volume of 25 µL, 1 X PCR buffer, 20 µM dNTP each, 0.2 µM primers each, 1.5 mM MgCl, 1U Taq polymerase (Promega) and 100ng DNA template. Reaction cycle was comprised of an initial step of denaturation for 5 minutes at 92ºC followed by 40 cycles each consisting of a denaturation step of 15 seconds at 92ºC, an annealing step 30 seconds at 52ºC and an extension step of 45 seconds at 72ºC. Ten minutes were given after the last cycle to the extension step at 72ºC to ensure the completion of reaction. PCR amplified products was analyzed on 1% agarose gel containing 2 µL of ethidium bromide (10 mg/mL) in 1X Tris-borate EDTA (TBE) buffer (Tris 108 g/L, borate 55g/L, and EDTA 7.5 g/L). Amplified samples were mixed with loading buffer (20% glycerol, 0.25% bromophenol blue) and was electrophoresed at 80V until the bromophenol blue migrated approximately two third of the length of the gel. DNA bands were observed in UV trans-illuminator and their photographs were taken.
Cluster analysis was carried out for reaction towards ELISA, tillers, height and grain weight.
Results
Evaluation of F2 Genotypes on the Basis of ELISA, Grain Weight, Tillers and Height
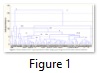
Finally all the selected 61 potential sources of resistance of F2 population were analyzed regarding their reaction towards ELISA, tillers, height and grain weight (Table. S1). Cluster analysis revealed two main groups A and B which were further divided into A1, A2, B1 and B2 (Fig. 1). Only one genotype i.e. 33 present in A2 group showed different behavior based on cluster analysis. The OD value recorded in this genotype was 0.403, 0.203 and 0.317 for PAV, MAV and RPV respectively while tillers counted were 7, height checked was 93 cm and grain weight was 4.40 gms. Subgroup A1 involved four genotypes 49, 96, 418 and 421 behave differently from others. Genotype 96 reacted negatively against all the three isolates having 2 tillers whereas height was 88 cm and exhibited 1.69 gms grain weights. While genotype 421 showed positive ELISA response, numbers of tillers were 8, height 77 cm and grain weight was 2.26 gms.
Group B was further categorized in to B1 and B2. Group B1 was divided into B1a and B1b. Subgroup B1a had four genotypes and all showed distinct behavior. Genotype 89 had ELISA value of 0.242 (PAV), 0.179 (MAV) and 0.173 (RPV) while tillers were 19, height record was 112 cm and grain weight measured was 4.28 gms whereas the ELISA values for PAV, MAV and RPV for genotype 81 were 0.171, 0.181 and 0.173 respectively, had 14 tillers, while height was 115 cm and exhibited 4.28 gms grain weight. Subgroup B1b was further sub divided into B1b(i) and B1b(ii). Subdivision B1b(i) had two classes (i)a and (i)b. Class (i)a involved 12 genotypes in which 412 and 50 behave differentially from others. Genotype 412 showed negative ELISA response against the three tested isolates, with 2 tillers, 98 cm height while grain weight was 3.78 gms. While genotype 50 having positive ELISA response against PAV, MAV and RPV, consisted of 8 tillers height was 102 and seed weight record was 3.62 gms. Two genotypes 48 and 53 were involved in class (i)b. Genotype 48 behave positively for ELISA whereas tillers counted for this genotype was 3, height measured was 96 cm and 4.40 gm grain weight. Subdivision B1b(ii) had two more classes i-e (ii)a and (ii)b. 20 genotypes fell in (ii)a class and they all showed different behavior. Genotype 56 reacted positively to BYDV-PAV but negative to BYDV-MAV and CYDV-RPV. The tillers of this genotype were 4; height was 107 cm and 4.25 gms grain weight. While genotype 68 behaved negatively to PAV and RPV but showed positive response to MAV, with 5 tillers 107 cm height and 3.83 gms grain weight whereas genotype 139 in class (ii)a showed resistance to BYDV-PAV and MAV but remained susceptible for CYDV- RPV, and exhibited 8 tillers, the height checked was 108 cm and grain weight record was 2.60 gms. Class (ii)b involved 17 genotypes and all showed different responses. Genotype 110 had low virus titer for PAV MAV and RPV, had 15 tillers while height 104 cm and having grain weight of 4.48 gms whereas genotype 142 react positively only MAV (0.226) and showed low OD for PAV (0.128) and RPV (0.200) having 10 tillers, 107 cm height and showing grain weight of 2.86 gms and had distinct behavior. Group B2 included only one genotype that showed distinct behavior from all other genotypes having ELISA values of 0.150 (PAV), 0.217 (MAV) and 0.196 (RPV), having 19 tillers, height of 107 cm and grain weight of 3.56 gms.
Evaluation of resistance on Visual Symptom Score (VSS)
Under field conditions, infected plants of oats showed typical symptoms of reddening and were also used as a source of BYDV inoculums. The 61 F2 genotypes were observed closely for the presence of symptoms of BYDV /CYDV at the phase of full flowering on 0-5 scale. Attack of BYDV /CYDV was associated with color changes of leaves and reduction in yield. Six (6) out of 61 genotypes did not show any symptoms of BYDV /CYDV and they exhibited visual symptom score 1 and these were 60, 91,110,119,121 and 129 .They were ranked as resistant. Whereas genotypes 53, 55, 56 and 57 had a VSS of 5 and showed leaf discoloration which is typical symptoms of BYDV and graded as highly susceptible. Thirty two (32) out of remaining 51 genotypes showed only moderate symptoms (MR) and considered as moderate resistant genotypes. The remaining 19 genotypes were remained moderately susceptible (MS). (Table S1)
Confirmation of Bdv2 Gene by SCAR Primers
DNA of 12 randomly selected F2 genotypes were tested with SCAR marker for the development of backcross. The SCAR primer BYAGIR/F could not amplify the band diagnostic for Bdv2 gene in our segregating population. Therefore new SCAR primers BCF5 and BCR6 were developed from the sequence of T. intermedium found in NCBI database. Presence of Bdv2 gene was confirmed in 6 out of 12 by using these primers. They amplified a fragment of 400bp representing the presence of Bdv2 gene (Table S1). The positive control TC14 also amplified product of 400bp conferring the presence of Bdv2 gene while negative control Inqilab91 did not amplified any product (Fig. 2). The six genotypes i.e. 91, 96, 110, 119, 121 and 131 were backcrossed with recurrent parent Inqilab91 and their seeds were collected.
Data & Tables
Discussion
Climate change is a burning issue since the start of 21st century. Besides, deteriorating earth abrupt climate change has direct or indirect impact on biotic and abiotic stresses suffered by crop plants. Keeping in view the increasing rate of population of the world and climate change-induced increase in temperature wheat production is expected to reduce by 2050 [29]. One of the biotic factors damaging the wheat is aphids. Aphids not only damage the crop by sucking the leaf sap but, they also transfer the BYDV /CYDVs into the plant cells. To mitigate the problem of aphid, farmers mostly used some insecticides at adult stage which work effectively but for viruses some inherited resistance is required. Bdv2 genes were well reported to have potential against BYDV /CYDVs. In the current perusal, the presence of resistance against CYDV /BYDV in F2 populations was evaluated during 2008. In the same year there was high incidence of yellow rust due to artificial inoculation in adjacent fields. Among seven F2 populations only three were resistant to rust and rests were highly susceptible. Therefore, the susceptible genotypes were discarded after field evaluation. The three F2 populations consisting of 61 genotypes in total were studied thoroughly for evaluation of resistance on the basis of symptoms, phenotypic data and expression analysis by ELISA. Though leaf discoloration was obvious to differentiate resistant and susceptible genotypes but BYDV symptoms do not represent the best criterion for selection of resistance or tolerance as symptom expression in field condition was highly variable. Variation in time of infection, temperature, plant density and tillering ability can influence the appearance and intensity of symptoms [27]. In connection to these findings selection and breeding only based on symptoms may be misleading. On the basis of combined effect of ELISA and agronomic values genotypes were placed in different categories i.e. susceptible, tolerant, moderately resistant and resistant. According to this classification, genotypes such as 49, 53, 55, 96, 98, 139, 142, 416, 418, 419 and 421 were considered as susceptible as they showed high ELISA values as well as low grain weight less tillers and reduced height. But it was also observed that genotypes 31, 47, 48, 50, 52, 56, 57, 60, 61, 94, 103, 106, 113, 115, 127,140 and 413 had moderate grain weight and had increased height but less tillers and also showed high optical density these were considered as tolerant in which even high virus titers cause negligible effect on yield which are in line with earlier reports [30]. Genotypes 62, 69, 93, 108, 109, 122, 126, 131, 132, 133, 146 and 411 showed moderate grain weight, increased height and tillers and low absorbance ratio hence, they were called moderately resistant. There were still other genotypes 33, 68, 77, 78, 90, 99, 105, 111, 112, 116, 119 and 129 which had low virus titer and grouped in the category of moderately resistant as they also had moderate grain weight, moderate height and tillers. Genotypes 30, 81, 89, 91,101, 110 and 121 were ranked as resistant as they had maximum grain weight, maximum tillers and good plant height and had low virus titer indicating the presence of Thinopyrum translocation that interfere with virus multiplication [20] or reduced cell to cell movement [21].
Generally the backcross is continued for five to six generations so that the resistant gene has been fully recovered in recurrent variety but due to short duration of study it was not possible to evaluate BC1F1 as well as to proceed for further backcrosses. Majority of F2 genotypes proved to be moderately resistant and resistant against BYDV /CYDV with good agronomic values. As cases for BYDV /CYDV are gradually increasing in Pakistan due to favorable environmental conditions and the presence of inoculum so there is need to develop resistant germplasm before any severe epidemic. The generated F2 germplasm offers good prospects for use in wheat improvement program carried out at CDRP-NARC.
References
- Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, et al. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annual Review of Phytopathology, (2011); 49465-481.
- Noorka IR, Shahid SA (2013) Use of conservation tillage system in semiarid region to ensure wheat food security in Pakistan. Developments in Soil Salinity Assessment and Reclamation: Springer. pp. 769-782.
- Oswald JW, Houston BR. Host range and epiphytology of the cereal yellow dwarf disease. Phytopathology, (1953); 43309-313.
- Wang MB, Abbott DC, Waterhouse PM. A single copy of a virus‐derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Molecular Plant Pathology, (2000); 1(6): 347-356.
- Khalid S, Aftab M, Ahmad I, Aslam M. Detection of barley yellow dwarf virus in Pakistan. Pakistan Journal of Botany, (1992); 24(2): 225-226.
- Miller and WA, Rasochová L. Barley yellow dwarf viruses. Annual Review of Phytopathology, (1997); 35(1): 167-190.
- Kundu J, Jarošová J, Gadiou S, Cervena G. Discrimination of three BYDV species by one-step RT-PCR-RFLP and sequence based methods in cereal plants from the Czech Republic. Cereal Research Communications, (2009); 37(4): 541-550.
- Kraakman A, Martinez F, Mussiraliev B, Van Eeuwijk F, Niks R. Linkage disequilibrium mapping of morphological, resistance, and other agronomically relevant traits in modern spring barley cultivars. Molecular Breeding, (2006); 17(1): 41-58.
- Ayala L, Van Ginkel M, Khairallah M, Keller B, Henry M. Expression of Thinopyrum intermedium-derived Barley yellow dwarf virus resistance in elite bread wheat backgrounds. Phytopathology, (2001); 91(1): 55-62.
- Goulart L, Ohm H, Foster J. Barley yellow dwarf symptom severity in oat affected by plant growth stage at infection and plot type. Crop Science, (1989); 29(6): 1412-1416.
- Yan W, Hunt L. Interpretation of genotype× environment interaction for winter wheat yield in Ontario. Crop Science, (2001); 41(1): 19-25.
- Makkouk KM, Kumari SG. Epidemiology and integrated management of persistently transmitted aphid-borne viruses of legume and cereal crops in West Asia and North Africa. Virus Research, (2009); 141(2): 209-218.
- Rasmusson DC, Schaller C. The inheritance of resistance in barley to the yellow-dwarf virus. Agronomy Journal, (1959); 51(11): 661-664.
- Sharma H, Gill B, Uyemoto J. High levels of resistance in Agropyron species to barley yellow dwarf and wheat streak mosaic viruses. Journal of Phytopathology, (1984); 110(2): 143-147.
- Brettell R, Banks P, Cauderon Y, Chen X, Cheng Z, et al. A single wheatgrass chromosome reduces the concentration of barley yellow dwarf virus in wheat. Annals of Applied Biology, (1988); 113(3): 599-603.
- Xin Z, Brettell R, Cheng Z, Waterhouse P, Appels R, et al. Characterization of a potential source of barley yellow dwarf virus resistance for wheat. Genome, (1988); 30(2): 250-257.
- Xin Z, Xu H, Chen X, Lin Z, Zhou G, et al. Development of common wheat germplasm resistant to barley yellow dwarf virus by biotechnology. Science in China Series B, Chemistry, Life Sciences & Earth Sciences, (1991); 34(9): 1055-1062.
- Banks P, Xu S, Wang R-C, Larkin P. Varying chromosome composition of 56-chromosome wheat× Thinopyrum partial amphiploids. Genome, (1993); 36(2): 207-215.
- Larkin P, Banks P, Lagudah E, Appels R, Xiao C, et al. Disomic Thinopyrum intermedium addition lines in wheat with barley yellow dwarf virus resistance and with rust resistances. Genome, (1995); 38(2): 385-394.
- Sharma H, Ohm H, Lister R, Foster J, Shukle R. Response of wheatgrasses and wheat× wheatgrass hybrids to barley yellow dwarf virus. Theoretical and applied genetics, (1989); 77(3): 369-374.
- Anderson JM, Bucholtz DL, Greene AE, Francki MG, Gray SM, et al. Characterization of wheatgrass-derived barley yellow dwarf virus resistance in a wheat alien chromosome substitution line. Phytopathology, (1998); 88(8): 851-855.
- Ayala L, Henry M, Gonzalez-de-Leon D, Van Ginkel M, Mujeeb-Kazi A, et al. A diagnostic molecular marker allowing the study of Th. intermedium-derived resistance to BYDV in bread wheat segregating populations. Theoretical and Applied Genetics, (2001); 102(6-7): 942-949.
- Stoutjesdijk P, Kammholz S, Kleven S, Matsay S, Banks P, et al. PCR-based molecular marker for the Bdv2 Thinopyrum intermedium source of barley yellow dwarf virus resistance in wheat. Crop and Pasture Science, (2001); 52(12): 1383-1388.
- Hohmann U, Busch W, Badaeva K, Friebe B, Gill BS. Molecular cytogenetic analysis of Agropyron chromatin specifying resistance to barley yellow dwarf virus in wheat. Genome, (1996); 39(2): 336-347.
- Saleem K, Hameed S, Ul-Haque I. Phylogenetic analysis of coat protein gene of BYDV-MAV strain from wheat. Archives of Phytopathology and Plant Protection, (2013); 46(14): 1747-1755.
- Zamurrad M, Hameed S, Saleem K, ul Haque I, Kausar S. Phylogenetic analysis of coat protein gene of CYDV-RPV strain from Wheat. Advancements in Life Sciences, (2014); 2(1): 16-22.
- Schaller C. The genetics of resistance to barley yellow dwarf virus in barley; 1984. pp. 93-99.
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JD. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. The Plant Journal, (1993); 3(3): 493-494.
- Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. Plos ONE, (2013); 8(6): e66428.
- Singh RP, Burnett PA, Albarran M, Rajaram S. Bdv1: a gene for tolerance to barley yellow dwarf virus in bread wheats. Crop Science, (1993); 33(2): 231-234.