![]()
Insight of Tp53 Mutations and their effect on Protein in Different Feline and Canine Neoplasms
Rashid Saif1*, Ezza Khan2, Arooj Azhar2, Shahnaz Choudhary2, Tanveer Hussain1, Masroor Ellahi Babar1, Ali Raza Awan3, Muhammad Tayyab3, Saeeda Zia4, Muhammad Wasim3
Adv. life sci., vol. 3, no. 2, pp. 42-50, February 2016
*- Corresponding Author: Rashid Saif (Email: rashid.saif@vu.edu.pk)
Authors' Affiliation
2- Kinnaird College for Women, Lahore – Pakistan
3- University of Veterinary and Animal Sciences, Lahore – Pakistan
4- National University of Computer and Emerging Sciences, Lahore – Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
Supplementary Data
References
Abstract
Background: Mutations in the Tp53 gene, a tumor suppressor gene, may cause dysfunction in growing cells and hinder the phenomenon of apoptosis, an alleged cause of tumorigenesis. It is involved in conservation of the genome and DNA repair, mutations of this gene may cause the damaged cells to grow continuously.
Methods: The type of molecular changes in Tp53 gene and their effects on physiochemical and structural properties of this protein in various Canine and Feline cancers were observed in this study by using online bioinformatics tools.
Results: Our results indicated that lymphomas and perianal adenocarcinomas (PAC) have the same mutation at c. 104, while mammary tumors and canine transmissible venereal tumor (CTVT) contain different mutations. Referring to changes in protein, synonymous mutations in granulomas were observed while certain mutations in squamous cell carcinoma (SCC) and head & neck tumors were detected in Canis familiaris. In Felis catus, the mutant protein was similar to wild type protein with exception of mutant 5 of mammary tumor, which had a deletion at the 287 amino acid position.
Conclusion: The insight gathered on the p53 mutant proteins in both species aided our understanding of the in-vivo fate of the p53 protein and its isoforms and the effects that morphological changes can have on the fate of cells. Furthermore, isolation of this protein may augment our understanding about the structural biology of these proteins.
Keywords: Bioinformatics tools, Tp53 mutants, Feline tumors, Canine tumors, TP53 protein mutants
Introduction
DNA can be affected by two types of mutagenic events, exogenous and endogenous mutations, which lead to the deregulation in the mitogenic program and can eventually cause cancer. Cancerous genes can be divided into two categories that are affected by mutations. One category consists of the proto-oncogenes, in which there is over expression of the product after mutation. The second type of genes is the anti-oncogenes, which act as tumor suppressors, in which mutation causes inactivation of both copies of the gene [1]. About two decades ago the Tp53 gene was discovered, and since then it has garnered much attention in the field of oncology. Researchers have identified the gene as a tumor suppressor gene after extensive analysis of its wild type protein [2]. The protein that is coded by this gene (named p53 protein) has its role in the conservation of the genetic stability and serves as an effective transcription factor [3].
Cellular responses such as DNA repair, apoptosis and cell cycle arrest are triggered by the p53 protein under conditions like hypoxia, nutritional depletion, DNA damage and stimulation of the oncogene. The p53 protein protects the genome by hindering the growth of damaged cells. Additional functions that are performed by this protein are cellular differentiation, senescence and DNA metabolism [4]. The p53 protein acts as a transcription factor and binds on specific sites on DNA. In case any damage occurs to the DNA, there is increase in p53 and it arrests the cell cycle at G1 phase allowing it to repair. If the damage is not fixed than cell apoptosis takes place. Mutations in the p53 protein produce altered proteins; these altered proteins in turn bind to normal wild type p53 proteins while forming tetra homodimers, thus disrupting function and ultimately resulting in tumorigenesis [5].
P53 mutations are also a chief cause of tumor formation in companion pets like cats and dogs. About half of the deaths of these pets are due to cancer above the age of 10 years, and mostly these tumors are malignant. 1-2% canine and feline cancers results in death of the animal [6]. Mammary tumors are reported as frequently occurring in Felis catus, which are caused by mutations in the Tp53 gene [7]. P53 mutations in Canis lupus lead to different malignancies such as osteosarcoma, thyroid cancer and mammary tumors. Also, about 6 percent of the canine malignant tumors are lymphoma [8] and canine transmissible venereal tumor (CTVT), which is found in both genders, can spread to other bodily tissues as well [9]. The present study investigates the role of p53 in tumor formation in pets. A better understanding of the protein was made by applying bioinformatics tools on canine and feline samples. Wild type and mutant proteins both were analysed. The Tp53 gene is located on E1p14 chromosome of Felis catus, where as in Canis lupus it is located on chromosome 5 [10]. Studying these mutations helps to diagnose cancer at an early stage in animals. These companion animals can serve as a disease model. The mutations and pathways can be studied which may help in prognosis in humans, due to a similar homology to the human version of the gene and analogous tumor biology [10].
Methods
Collection of Samples
The wild type protein sequences of Tp53 gene in Canis lupis familiaris and Felis catus were taken from Ensemble Genome Browser (http://asia.ensembl.org/Canis_familiaris/Transcript/Summary?t=ENSCAFT00000026465&db=core) (http://asia.ensembl.org/Felis_catus/Transcript/Summary?t=ENSFCAT00000009625&db=core) whereas collection of tumor samples was done from the Pet Centre, University of Veterinary and Animal Sciences (UVAS) Lahore, Pakistan. The tumor samples included twenty dog tumor samples, aged 2-12 years of which five were mammary tumor samples, five were canine transmissible venereal tumor (CTVT) samples, three were perineal adenocarcinoma (PAC) samples, three were Squamous cell carcinoma (SCC) samples, two were granuloma samples and two were lymphoma samples. Six mammary tumor samples of Siamese breed cat, aged 2-7 years were collected. Core tissues of tumorous masses were taken for DNA extraction.
Primer Designing
Primers were designed using the wild type dog Tp53 sequence from ensemble genome browser (http://asia.ensembl.org/Canis_familiaris/Transcript/Summary?t=ENSCAFT00000026465&db=core) and the cat Tp53 sequence from ensemble genome browser (http://asia.ensembl.org/Felis_catus/Transcript/Summary?t=ENSFCAT00000009625&db=core) using Primer3 (http://primer3.ut.ee/) and Net Primer (http://www.premierbiosoft.com/netprimer/) software. The annealing time was 45 seconds and Tm used for primer designing was 60oC. A single set of primers was designed in each species for long range PCR. Forward and reverse primers in dog are 5’ CCCTGGTATAATGTTGCTGGAAG 3’ and 3’ GGATGAGGAAGGAGGTGACGTT 5’ whereas forward and reverse primers in cat are 5’ GGTACCTTTGTGTCTGGAGGACAT 3’ and 3’ GTACAGGTATGCCTCGGAAACAC 5’. Moreover, 7 and 3 internal sequencing primers were also designed in both species respectively.
DNA Extraction, PCR and Sequencing
DNA was extracted from the tumorous tissues of dog and cat using TIANamp blood DNA kit (0.1- 1 mL) (DP318-02), TIANGEN Biotech (Beijing) company. PCR amplification was done for sequencing. After sequencing of the PCR products, they were aligned using Sequencher Software (www.genecodes.com/).
Bioinformatics Tools
The protein sequences of mutants were generated from DNA sequences using Sequence Translation EMBL-EBI Tool (http://www.ebi.ac.uk/Tools/st/). Online bioinformatics tools were applied on the wild and mutant type protein sequences of Tp53 in Canis familiaris and Felis catus to analyse their physiochemical properties, secondary structures, conserved domains, transmembrane structures and post translational modifications. Protein sequences of wild and mutant types in both species were analysed with BLAST on NCBI (http://blast.ncbi.nlm.nih.gov/blast/Blast) before applying the following tools.
Physiochemical Properties
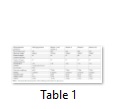
Protparam tool (http://web.expasy.org/protparam/) was applied for the analysis of physiochemical properties, composition of amino acids, molecular weight, atomic composition, theoretical PI, instability and aliphatic index, extinction co-efficient, GRAVY and estimated half-life in the protein sequences of wild and mutant type Tp53 in dog and cat (Table 1).
Secondary Structure Prognosis
Secondary structure of wild and mutant type protein sequences of Tp53 in both species was analysed using Psipred tool (http://bioinf.cs.ucl.ac.uk/psipred/). Bio Serf is another feature of PsiPred that provides the automated homology modelling [11]. A PDB file was saved for all the mutants and wild type and the structure was observed in Visual molecular dynamics (VMD).
Conserved Domains
Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan), NCBI CD Server (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) or Interproscan 5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) tool can be used for the analysis of conserved regions in the protein sequences. Motif Scan tool was used in the current study to analyse the conserved regions in the protein sequences of Tp53 in wild and mutant types in Canis lupis familiaris and Felis catus.
Transmembrane Structures
Protscale tool (http://web.expasy.org/protscale/) was applied to determine the transmembrane structures of the wild and mutant type protein sequences in both species. Kyte & Doolittle algorithm (http://web.expasy.org/protscale/pscale/Hphob.Doolittle.html), with threshold value above 1.6, was used during analysis. In order to get transmembrane structures window size was set to 19, whereas default window size of 9 in Protscale tool gives surface protein structures in the graph. Hydrophobicity, hydrophilicity and trans-membrane α- helices of the amino acids of the protein sequences of wild and mutant types of Tp53 in both species were analysed.
Post-translational Modifications
During translation the nascent polypeptides become functional after post-translational changes; they attach to various metals, prosthetic groups and organelles for post translational modifications [12]. Scanprosite tool (http://prosite.expasy.org/scanprosite/) was applied to study the post-translational changes like phosphorylation, n- myristoylation etc. in the protein sequences of wild and mutant types of Tp53 in Canis familiaris and Felis catus.
Results
Canine Tp53 protein analysis
Physiological properties
To analyse the physiochemical properties of the wild type and mutants was performed using protparam tool [13]. Mutant 1 lymphoma (M1), mutant 2 mammary tumor (M2), mutant 3 CTVT (M3), mutant 4 PAC (M4), mutant 5 SCC (M5), mutant 6 granuloma (M6) and mutant 7 head and neck tumor (M7) were observed. The results indicate the number of amino acids (a.a) in wild type is less and all mutants consisted of 689 a.a. The molecular weight is determined by the addition of water molecules and the results indicated that the wild type and mutants had different values; the no. of positive and negatively charged residues in wild type is 46 but in M1 and M2 the value was different. M3, M5, M6 and M7 showed similar results. The PI indicates the pH of the protein by using the pKa and proteins are compact and stable at PI, the wild type value is 7.25 however M1, M2, M4 and M3, M5, M6, M7 showed similar values. Formula and no. of atoms were given for the wild type but because of some ambiguous position in the sequence the atomic composition was not generated for the mutants. The “Extinction Coefficient” explains the amount of light that is absorbed by the protein and the value for all the mutants is same. The half-life of a protein specifies the time when the protein is degraded into half after its synthesis which for wild type is 4.4 hours and for mutants is 1.9 hours. Instability index describes stability of protein in test tube, if value is above 40 than protein is unstable and results indicate that wild type and mutant protein is unstable. The “Aliphatic Index” indicates the volume occupied by the aliphatic side chains and the value for M1, M4, M5, M6 and M7 is same. “GRAVY” indicates the hydrophobicity of the protein; more positive score indicates more hydrophobicity, results show negative value [14].
The secondary structure of the protein were observed using PsiPred [15]. The results showed similar secondary structure for M1 and M4, however M2 also had similar secondary structure though the mutation in M2 was different, helical structure is present from 13-18 and 67-76 a.a (Figure 1 in Sup. Data). In M3, helices are present at 12-20, 24-26, and 461-463 a.a. In M5, initially only one helix is present from 13-17 and 67-75 a.a (Figure 1c in Sup. Data). A cylinder represents helix, an arrow represents a strand and a straight line represents coil structure.
The protein structure of wild type and mutants were generated from Bioserf modelling. The wild type results indicate 3075 bonds, M1 and M4 show 5567 bonds and 689 residues, M2 showed 5564 bonds and 689 residues, M3 showed 5558 bonds and 689 residues, M5, M6, M7 showed 5560 bonds and 689 residues (Figure 2 in Sup. Data).
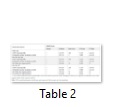
The motif scan tool was applied on the sequence of wild and mutant types of dog Tp53 protein sequences and the results of conserved domains obtained were analysed. Match details and match score were obtained which gave the status, position, raw score, N-score and E- value of the wild and mutant protein sequences (Table 2). Bipartite nuclear localization signal profile and proline rich region profile of the wild type, M1, M2, M3 and M4 are the same (Figure 3a, c in Sup. Data). M5, M6 and M7 have two proline rich regions and different signal profiles (Figure 3b, d, e in Sup. Data). Major vault protein (MVP) repeats profile is same in the wild and mutant types of protein sequences as shown in (Figure 3f in Sup. Data).
Transmembrane structures of the wild and mutant type protein sequences were observed using protscale tool. We used the Kyte & Doolittle algorithm, which has threshold value above 1.6. The protein sequence of wild type has two transmembrane structures (Figure 4a in Sup. Data), M1 and M4 (Figure 4b in Sup. Data), M2 (Figure 4c in Sup. Data), M5, M6 and M7 (Figure 4e in Sup. Data) have five transmembrane structures whereas M3 showed six transmembrane structures (Figure 4d in Sup. Data). Post translational modifications of the wild and mutants of the protein Tp53 in Canis lupus are shown in Table 3. “[ ]” in the consensus sequence of the post translational sites shows that any one of the amino acids mentioned in this bracket may be present at this position, while an amino acid within“{}”shows that there can be any amino acid at that site except for the one indicated within the braces. Similarly, parenthesis “( )” shows the number of amino acids that should be present at a particular site and “x” shows any of the amino acids.
Feline Tp53 protein analysis
Physiological properties
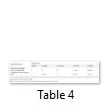
Protparam was used to assess the physiochemical properties of the feline protein. The results indicate, no. of amino acids were 386, molecular weight was 42692.4, the theoretical PI was 7.12, no. of positive and negative residues were 46, the formula was C1870H2944N530O571S22, the no. of atoms were 5937, extinction coefficient was 34670, half-life was 30 hours, instability index was 69.36 which indicates instability, the aliphatic index was 63.70 and GRAVY was -0.621 which indicates less hydrophobicity. The results of M5 indicate that the no. of amino acids are 385 (one amino acid less as compared to the wild type and other mutants), the molecular weight was 42563.3, PI was 7.55, the no. of positive and negative charged residues were 45 and 46, the formula was C1865H293N529O568S22, the no. of atoms were 5921, the extinction coefficient was 34670 which is similar to wild type, half-life was 30 hours, instability index was 69.21 which means it is unstable, aliphatic index was 63.87 and GRAVY was -0.614 which means it is less hydrophobic. There is only slight difference between the wild type and M5, as there is difference of only 1 amino acid. The secondary structures of wild type and M5 were observed by using the PsiPred tool. In M5 there was a change in the strand as it was elongated from position 101-107 and then from 114-117 (Figure 5 in Sup. Data). BioSerf was used for Felis catus to obtain the structure of mutants and wild type, the results indicated that the wild type and M1, M2, M3, M4 have the same results of 3075 bonds and 386 residues. In M5 there are mutations in the protein so differences were found that is 3066 bonds and 385 residues (Figure 6 in Sup. Data). Conserved domains in the Tp53 protein sequences of wild and mutant types of Felis catus were also observed using motif scan. The conserved regions contain few amino acid enriched regions and certain domains as well, which in the protein sequence of wild and mutant types of cat is bacterial immunoglobulin like domain-1. Results of match score indicate that the Big-1 domain profile (Figure 7a in Sup. Data) and bipartite localization signal profile (Figure 7b in Sup. Data) present in the protein sequences of wild and mutant types are the same (Table 4). Local alignment of the two protein sequences showing the conserved domains is shown in (Figure 7c in Sup. Data). In order to find transmembrane structures of wild and mutant types, the Kyte & Doolittle algorithm was set which gave three transmembrane structures of wild and mutant protein sequences (Figure 8a, b in Sup. Data).
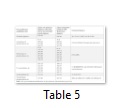
Analysis of the post translational changes in the wild and mutant type protein sequences of Tp53 in Felis catus was carried out using Scanprosite. Changes such as Tp53 family signature, CK- II phosphorylation site, protein kinase C phosphorylation site, n- myristoylation site, n-glycosylation site, tyrosine kinase phosphorylation site were observed as shown in Table 5.
Data and Tables
Discussion
The Tp53 gene was studied in twenty samples of canine tumors. The wild type sequence was used as the reference sequence and the mutations in the samples were studied. Multiple sequence alignment (MSA) was performed using Sequencher software and mutations were analysed. Major mutations in exon 3 were seen in the results, whereas fewer mutations in exon 4 and 5 were observed. In lymphoma at c.104 “G” is replaced by “A”. In mammary tumor at c.76 “C” is replaced by “T” in most samples, whereas in some samples it is replaced by “G”. At c.87 C>A, at c.90 and c.92 “T” is replaced by “G” and at c.356 A>C, in CTVT, at c.76 C>T whereas in some samples it is replaced by “G” and at c.90, c.92, c.94 T>G. In PAC at c.87 C>A and at c.104 “G” is replaced by “A”. In SCC, granuloma and Head & Neck tumor, synonymous mutations were observed. Protein translation of CDS was done and mutant proteins were analysed. The wild type protein consisted of 383 amino acids but the mutant protein consisted of 689 amino acids thus an entirely abnormal protein was formed due to frameshift. BLAST was used to identify the similarities in mutant protein. The results indicate that M1 and M4 in dog showed similar mutations, at position 35 Glutamine was present. In M2 at position 30 with respect to M1 Glutamic acid was present and at position 35 Arginine was present. In M3 at position 27 and 32 Valine was present and at position 35 Arginine was present. M5, M6, M7 showed similar mutations; at position 35 Arginine was present.
The physiochemical properties showed the increase of amino acids in mutants because of abnormal protein formation as a result the molecular weight of the mutant protein increased from the wild type. The positive and negative charged residues of mutants are higher than the wild type. The extinction coefficient was elevated in mutant that means that the absorbance capacity of mutant is more than the wild type. The half-life of the mutant protein l.9 and 4.4 hours for the wild type respectively. Proteins having an in vivo half-life less than 5 hours have instability index above 40, that in unstable [16]. The GRAVY values are in negative so it means that the hydrophobic character is reduced however the mutant values less so it is less hydrophilic than wild type. PsiPred results indicate the amino acid sequence of the mutants and wild type along with confidence value that has a range from 0-9. M1, M2, M3 showed the same results even after a different mutation without any structural change [17]. BioSerf uses Psipred algorithm as a template and then generates a model for the protein by using modeller to generate appropriate model [18]. Motif scan tool showed conserved regions in the protein sequences of wild and mutant types in dog. Conserved domains were initially considered as autonomously or stable folding units in a protein that have three dimensional structure [19]. The domain is highly conserved in case of the more elevated bars, the white bars indicate particular amino acid is conserved but not on the exact site in a particular domain whereas green bars specify that the particular amino acid is conserved in all domains and is present at the correct place (Figure 3 in Sup. Data). Waterman Smith algorithm score was observed to be 122 which is directly related to the conserved domains. In case of high score, domain is more conserved and vice versa. Conserved domains contain certain regions in a protein sequence, enriched with one or more specific amino acids, which is proline rich region in this case. MVP in the conserved domain is a component of vault which is a complex of ribonucleoproteins, performing diverse functions like signal transduction, transport and multidrug resistance mechanisms [20]. The structure of vault is globular made up of MVP having two similar fractions, consisting of 39 copies of MVP. The moieties of vault can be dissociated at acidic pH [20]. N-terminal of MVP has 8 copies of vault tandem repeats and C terminal is involved in oligomerization activity (http://prosite.expasy.org/PDOC51224). Transmembrane proteins are polytopic in nature which form precipitates and aggregates in water and belong to the integral class of proteins. They have two forms which are α helices present in inner and outer membranes of the cell whereas β barrels are only present in the outer membranes. The information obtained via Scanprosite tool verified that Aspartate and glutamate are acidic in nature, present at three residues away from the C terminal of the phosphate acceptor site in CK- 2 reactions. Phosphorylation rate is decreased if the basic residue is present at the N- terminal and increased if the acidic residue is present (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00006). The Vmax and Km of the phosphorylation reaction is increased if the basic residue is present at the N or C terminal and the protein kinase C phosphorylates only serine or threonine residue (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00005). In glycosylation reactions, if proline amino acid is present between Asn and Ser or Thr residue than the glycosylation is inhibited (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00001). Asp or Glu are the acidic residues for phosphorylation in tyrosine kinase reactions and are present at N terminal along with lysine and arginine which are substrates of the reaction (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00007). Amidation reaction can be carried out with all amino acids but the Val and Phe which are hydrophobic neutral residues are good substrates in comparison with charged residues like Asp and Arg (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00009).
Mammary tumor was studied in Felis catus. Mutant 1-5 showed mutations in the exon 3, 4, 7 and 9. M1 showed mutation at c.105 A>G, at c. 465 “T” was replaced by “C”, at c.1050 G >A. In M2 at c.105 and at c.1050 “G” was replaced by “A”. In M3 at c.1050 G>A, in M4 at c.105 G>A and in M5 at c.465 “T” was replaced by “C” and at c.895 G>T. The protein translation was performed of all the mutants and wild type and Blastp was executed. Blastp results signified that even after mutations in the CDS there were no mutations in the protein of M1, M2, M3, M4 and showed same results with wild type protein, however in M5, mutations in the protein was found. The secondary structure indicated by PsiPred shows that M5 consisted of more extended structure as strands are elongated. The BioSerf model confirmation specifies that there are 386 residues in wild type and 385 residues in M5 so the difference of 1 a.a is confirmed, further the wild type consists 9 more bonds than M5.
In the conserved regions of wild and mutant protein sequences of cat bacterial immunoglobulin like domain- 1 is present (Fig, 7a). Big-1 domain is present in adhesion molecules of bacteria which play role in pathogenicity. The name Big-1 is given to the domain because of the three dimensional structure of the domain in certain adhesions like Yersinia pseudotuberculosis invasin and enteropathogenic Escherichia coli intimin (http://prosite.expasy.org/PDOC51127). In post translational modification transformed cells and non-transformed proliferating cells have p53 tumor antigen protein in very large amounts as compared to resting cells having very low amounts of p53 protein. P53 protein is composed of nearly 390 amino acids and belongs to a family of phospho proteins. Excessive post translational modifications are observed in case of tp53 activation by genotoxic stresses leading to heavy phosphorylation of the N terminal residues [19, 21]. These amino acids of the protein can be further divided into four domains which include, hydrophobic proline-rich region at position 80-150, central domain region from 150-300, highly charged acidic domain of almost 75-80 residues, and highly basic C-terminal residue. The central region domain was selected as a p53 family signature which has 13 residues and is a main domain which is involved in point mutations in tumors (http://prosite.expasy.org/cgi-bin/prosite/nicedoc.pl?PS00348). This study revealed various characteristics of Tp53 protein in feline and canine tumors and their comparison with their wild type protein sequences. The hotspot mutation region was exon 3. Mutations in the canine tumors resulted in abnormal protein formation and thus causing tumor formation. The results obtained via different tools of bioinformatics showed that protein sequences of all the mutants in dog vary from each other due to mutations. Prominent mutations in CTVT and mammary tumor were unveiled in dog. On the other hand, no changes in the protein sequence were observed despite the mutations in cat mammary tumor CDS, except in M5, which has undergone mutation in single amino acid.
References
- Fromentel CCD, Soussi T. TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes, Chromosomes and Cancer, (1992); 4(1): 1-15.
- Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature, (1989); 342(6250): 705-708.
- Liu J, Zhang C, Feng Z. Tumor suppressor p53 and its gain-of-function mutants in cancer. Acta Biochimica et Biophysica Sinica, (2014); 46(3): 170-179.
- Bellini MF, Cadamuro ACT, Succi M, Proença MA, Silva AE. Alterations of the TP53 gene in gastric and esophageal carcinogenesis. BioMed Research International, (2012); 2012.
- Moro J, Tinucci-Costa M, Silveira A, Gerardi D, Alessi Reactivity of p53 protein in canine transmissible venereal tumor. Arquivo Brasileiro de Medicina Veterinária e Zootecnia, (2010); 62(2): 318-323.
- Gentschev I, Patil SS, Petrov I, Cappello J, Adelfinger M, et al. Oncolytic virotherapy of canine and feline cancer. Viruses, (2014); 6(5): 2122-2137.
- Pang L, Blacking T, Else R, Sherman A, Sang H, et al. Feline mammary carcinoma stem cells are tumorigenic, radioresistant, chemoresistant and defective in activation of the ATM/p53 DNA damage pathway. The Veterinary Journal, (2013); 196(3): 414-423.
- Veldhoen N, Stewart J, Brown R, Milner J. Mutations of the p53 gene in canine lymphoma and evidence for germ line p53 mutations in the dog. Oncogene, (1998); 16(2).
- Stockmann D, Ferrari H, Andrade A, Cardoso T, Luvizotto M. Detection of the tumour suppressor gene TP53 and expression of p53, Bcl‐2 and p63 proteins in canine transmissible venereal tumour. Veterinary and Comparative Oncology, (2011); 9(4): 251-259.
- Cho K-W, Okuda M, Endo Y, Satoh H, Kang C-B, et al. Assignment of the cat p53 tumor suppressor gene (TP53) to cat chromosome E1p14→ p13 by fluorescence in situ hybridization. Cytogenetic and Genome Research, (1997); 79(1-2): 145-146.
- Buchan DW, Ward S, Lobley AE, Nugent T, Bryson K, et al. Protein annotation and modelling servers at University College London. Nucleic Acids Research, (2010); gkq427.
- Beevers L (1982) Post-translational modifications. Nucleic Acids and Proteins in Plants I. Berlin, Germany: Springer. pp. 136-168.
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Research, (2003); 31(13): 3784-3788.
- Ashokan K, Pillai M. In silico characterization of silk fibroin protein using computational tools and servers. Asian Journal of Experimental Science, (2008); 22265-274.
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics, (2000); 16(4): 404-405.
- Idicula‐Thomas S, Balaji PV. Understanding the relationship between the primary structure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Science, (2005); 14(3): 582-592.
- Yang J-Y, Peng Z-L, Chen X. Prediction of protein structural classes for low-homology sequences based on predicted secondary structure. BMC Bioinformatics, (2010); 11(1): 1.
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Research, (2013); 41(W1): W349-W357.
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Research, (2012); gks1243.
- Querol‐Audí J, Casañas A, Usón I, Luque D, Castón JR, et al. The mechanism of vault opening from the high resolution structure of the N‐terminal repeats of MVP. The EMBO journal, (2009); 28(21): 3450-3457.
- Appella E, Anderson CW. Post‐translational modifications and activation of p53 by genotoxic stresses. European Journal of Biochemistry, (2001); 268(10): 2764-2772.