Short Communication
Anticancer screening of medicinal plant phytochemicals against Cyclin-Dependent Kinase-2 (CDK2): An in-silico approach
Wajahat Khan1, Usman Ali Ashfaq1, Sadia Aslam2, Sidra Saif3, Tehzeeb Aslam3, Kishver Tusleem3,
Arooma Maryam4, Muhammad Tahir ul Qamar*5
Adv. life sci., vol. 4, no. 4, pp. 113-119, August 2017
*- Corresponding Author: Muhammad Tahir ul Qamar (Email: m.tahirulqamar@webmail.hzau.edu.cn)
Authors' Affiliations
2- Allama Iqbal Medical College Lahore – Pakistan
3- Fatima Jinnah Medical University, Lahore – Pakistan
4- Department of Biosciences, COMSATS Institute of Information Technology (CIIT), Islamabad – Pakistan
6- College of Informatics, Huazhong Agricultural University (HZAU), Wuhan – P.R. China
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Cyclin-Dependent Kinase-2 (CDK2) is a member of serine/threonine protein kinases family and plays an important role in regulation of various eukaryotic cell division events. Over-expression of CDK2 during cell cycle may lead to several cellular functional aberrations including diverse types of cancers (lung cancer, primary colorectal carcinoma, ovarian cancer, melanoma and pancreatic carcinoma) in humans. Medicinal plants phytochemicals which have anticancer potential can be used as an alternative drug resource.
Methods: This study was designed to find out anticancer phytochemicals from medicinal plants which could inhibit CDK2 with the help of molecular docking technique. Molecular Operating Environment (MOE v2009) software was used to dock 2300 phytochemicals in this study.
Results: The outcome of this study shows that four phytochemicals Kushenol T, Remangiflavanone B, Neocalyxins A and Elenoside showed the lowest S-score (-17.83, -17.57, -17.26, -17.17 respectively) and binds strongly with all eight active residues Tyr15, Lys33, Ileu52, Lys56, Leu78, phe80, Asp145 and Phe146 of CDK2 binding site. These phytochemicals could successfully inhibit the CDK2.
Conclusion: These phytochemicals can be considered as potential anticancer agents and used in drug development against CDK2. We anticipate that this study would pave way for phytochemical based novel small molecules as more efficacious and selective anti-cancer therapeutic compounds.
Key words: Cell cycle, CDK2, Cancer, Phytochemicals, Molecular docking
Introduction
In multicellular organism cell cycle is a synchronized chain of events directed by number of intracellular and extracellular signaling proteins which operate to maintain the fidelity of DNA content [1]. Central to cell cycle specific signaling protein repertoire are Cyclin Dependent Kinases (CDKs) complexed with cyclins. Cell cycle encompasses interphase and Mitotic phase, where interphase is morphologically categorized in G1, S and G2 phase. CDKs-Cyclin complexes act at check points during progression of events in cell cycle. They specifically guard the DNA content of cell and decide fate of cell to further move in cell cycle or not. CDKs belong to family of serine/threonine kinases that are activated at certain stages of interphase [2]. Activity of CDKs is regulated through a pattern of phosphorylation and dephosphorylation at conserved serine/threonine residues by proteins i.e. p40m015 as well as its binding with cyclin box on its partner cyclin [3]. In cell cycle progression decision, by far six mammalian CDKs complexed with 13 eukaryotic cyclin proteins at different events have been identified. In interphase, G1 phase characterized as phase of gap in which cell prepare itself for growth while CDK4, CDK5 and CDK6 in complex with cyclin D ensures the correctness of genetic content. Transition from G1 to synthesis (S) phase is critically regulated by CDK2-cyclin E complex. At this checkpoint, in case of any DNA content infidelity in cell, progression will be arrested to enter in S phase and p53 gene is activated for DNA repair. Once cell reaches in S phase, CDK2-cyclin A complex maintains a critical check on cells to let the healthy cell win the battle of survival within the human body [4]. Multiple cancer i.e. lungs cancer, primary colorectal carcinoma, ovarian cancer, melanoma, and pancreatic carcinoma have been reported due to over-expression or down regulation of CDK2 [5].
Asia accounts for 56% of total world’s population, add 44% to total global burden of cancer and contributes 51% to overall death toll globally [6-9]. CDKs are considered deliberate anticancer drug target to limit replicative potential of tumor cells. Therefore, small molecule CDK2 inhibition can play an important role in controlling the cancer and become a potential target for cancer therapy [10]. Therapeutic interventions of CDK2 inhibitors has been extensively studied in cancer and other proliferative diseases. Intensive research has led to discovery of 21 inhibitors that are known to block the activity of 11 isoforms of CDKs. Approximately 20 of them are in clinical trials. AG-024322 (Pfizer), AT7519 (Astex) and alvocidib another name of flavopiridol (sanofi-Aventis) are CDK2 specific inhibitors currently in pre-clinical studies. Recently in February 2015, palbociclib (Pfizer, Ibrane®), a CDK4, CDK6 inhibitor has been launched in market after Food and Drug Administration (FDA) approval [11-13]. For CDK2 inhibitors clinical data reports multitarget potential which negatively affect interconnected signaling pathway and produce undesired side effects. To discover specific CDK2 inhibitors with desired activity in addition to minimal effect on downstream signalosome and undesired effects, we need to direct our research on screening and development of phytochemical based small molecules library screening [5,14].
Medicinal plants are the natural source of therapeutic agents. A remarkable number of modern drugs have been isolated from plants based on their traditional use. These are also considered to be useful in eradicating the adverse effects of various chemotherapeutic agents as well as in prolonging longevity and attaining positive general health [15]. They produce different secondary metabolites generally called phytochemicals (flavonoids, alkaloids, terpenoids, polyphenolics etc.) which have anticancer potential and can be used as an alternative cancer drug resources [16,17]. Therefore, this study was designed to find out anticancer potential of phytochemicals which could target CDK2 and inhibit its over-expression by using in-silico methods (molecular docking and drug scan). The result of this study will facilitate the researchers to improve the status of anticancer therapeutics by diversifying the scaffolds of CDKs inhibitors.
Methods
Structure based virtual screening strategy was applied through high performing computing work station with following specifications (Intel(R) Core(TM) i5-3210M CPU @ 2.50 GHz, 5 Core(s) processor with 4.00 GB RAM and 64-bit Windows-8 Operating System). Structure based drug screening was performed with Molecular Operating Environment (MOE v2009) [18].
Ligand database preparation and receptor protein refinement
Structures of phytochemicals were retrieved from MPD3 database [19], MAPS database [20], ZINC database [21] and PubChem [22]. Ligand library composed of 2300 compounds was optimized for docking. Optimization involves addition of partial charges and energy minimization of selected compound through Protonate-3D and MMFF94X force field respectively. Optimized ligands files were stored in ligand database which was used later as an input file for docking studies.
CDK2 structure refinement
Likewise, three-dimensional structure of CDK2 was retrieved from the Protein Data Bank (PDB) using PDB ID: 3PXF with 1.8Å resolution [23]. To refine the protein structure, already bound ligands and water molecules were removed from the structure, 3D protonation and energy minimization was done in MOE. The minimized structure was used for docking in subsequent steps.
Molecular docking
MOE docking tool was used to dock phytochemical ligand library against allosteric ligand binding site of CDK2. Potential binding pocket containing the reported residue (Lys56) was identified with the help of site finder tool of MOE and was further taken through docking process. Ten best docked poses were generated through applying a scoring function London dG. Refinement of docking procedure was done by applying forcefield algorithm which keeps the receptor rigid. From them, best interacting ligands, molecules were screened on the basis of RMSD (Root-Mean-Square Deviation) which is usually measured in Angstrom (Å), and docking score. Already reported co-crystallized ligand, Fluorophore-8-anilino-1 naphthalene sulfonate (ANS) was used as a reference compound for docking into native CDK2 pocket to validate our docking result. Ligand receptor interaction analysis was done through LigX tool in MOE. It is designed to show potential residues interacting with the ligand molecules graphically. It generates 2D images representing the forces stabilizing ligand molecules within binding pockets of receptors.
Drug scan
To analyze the drug likeliness of screened phytochemical based lead molecules drug scan of final selected phytochemical hits was performed in MOE by ligand properties checking tool. Compounds were further filtered on the basis of the bearing appropriate molecular properties to be a drug candidate.
Results
Analysis of allosteric binding of 2300 phytochemical based small molecules to CDK2 receptor initially revealed 10 compounds which fit well within the binding pockets with low S-score. Iterative docking of the best docked 10 molecules within receptor pocket, originally occupied by co crystallized inhibitor Fluorophore-8-anilino-1 naphthalene sulfonate (ANS), resulted in 4 best phytochemicals with dock score comparatively less than the original one.
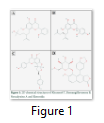
Chemical structure of our four selected phytochemicals; Khusenol T, Remangiflavanone B, Neocalyxins A and Elenoside are shown in Figure 1. Khusenol T (Fig 1A) is an essentail oil compund (C15H24O) isolated from Angola vetiver [20], Remangiflavanone B (Fig1 B) is a tetrahydroxyflavanone aromatic compound (C25H28O6) isolated from Physena madagascariensis [24], Neocalyxins A (Fig1 C) is a diarylheptanoids extracted from Alpinia blepharocalyx seeds [25] and Elenoside (Fig1 D), commonly known as arylnapthalene lignan is extracted from Justicia hyssopifolia [26].
Binding affinity analysis of selected compounds through LigX shown in Figure 2, revealed that top ranked molecules experience water mediated binding with the crucial catalytic residues of pocket.
Spatial position of Kushenol T is stabalized within the pocket (Figure 2A) with the lowest S-score (-17.83) and interacting through hydrogen bonds with Tyr15 and Phe146 while rest of close lying residues (Lys33, Ileu52, Lys56, Leu78, Phe80, Asp145) are having weak electrstatic interactions with Kushenol T. Following this is Remangiflavanone B inlaid within the same binding pocket as shown in Figure 2B, lying closer to Tyr15, Lys33, Ileu52, Leu55, Lys56, Leu66, Leu76, Leu78, phe80, Asp145 and Phe146 residues of CDK2 binding pocket with dock score -17.57 kcal/mol. Third best screened compound Neocalyxins A experiences water mediated and hydrogen bonding with Tyr15, Lys33, Asp145 and Phe146 while other closely lying residues are Ileu52, Leu55, Lys56, Val64, Leu66, Leu78 and Phe8o shown in Figure 2C. Elenoside has been shown to in laid well within the binding pocket showing water mediated interaction with Leu55, Lys56, in addition to non-covalent interactions with Val64 (Figure 2D). While calcualted RMSD value of all the four selected inhibitors with respecct to their co-crytallized CDK2 Fluorophore-8-anilino-1 naphthalene sulfonate (ANS) complex is given in Table 1, while binding conformation of each ligand inside the binding pocket of CDK2 is represented in Figure 3.
Drug scan
To verify the drug ability of selected phytochemicals, ligand properties were calculated with LigX tool of MOE. All selected phytochemicals showed positive results and fulfill the criteria of the Lipinski’s rule of five [27]. The rule describes that potential drug like compound should not have more than 5 hydrogen bond donors, maximum 10 hydrogen bond acceptors, molecular mass less than 500 daltons, and an octanol-water partition coefficient log P not greater than 5. Drug scan results are given in shown in Table 1.
Tables & Figures
Discussion
In eukaryotes, cell’s count is critically regulated by the release of growth promoting factors that dictate the entry of cell into subsequent cycles of Cell cycle. Cancerous cells lose their homeostatic control over growth promoting signal which render them as master of their own fate [28]. Cyclin-dependent kinases (CDKs) are serine/threonine protein kinases that are intimately engaged in regulating cell division cycle. Six CDKs have been identified so far in mammal, each one is required to make complex with cyclins at different stages of cell cycle. Cyclin-CDKs complex functions as a cell cycle checkpoint which arrest the cell cycle progression in case of DNA damage. Activity of CDKs is governed by its complex formation with Cyclin through 100 amino acid long cyclin box. Cyclin-Dependent kinase-2 (CDK2) complexed to Cyclin E and cyclin A at G/S junction and S phase of cell cycle clock respectively are important for continued replication of DNA, DNA damage repair phosphorylation and transcription of necessary growth factors [29]. Normal cells acquire the hallmarks of cancer and attain limitless cell replication potential. To inhibit the process of carcinogenesis, CDKs-cyclin complexes are attractive anti therapeutics targets. To cope with the constant need of novel and effective small molecule, anti-cancer therapeutics with minimal side effects, research is now focusing more on computational drug discovery [16]. Multiple studies reported anticancer potential of phytochemicals through computer aided drug design [30-32].
Despite of efforts spanning over 20 years only one FDA approved CDK2 inhibitor is available in market while rest of discovered CDK2 inhibitors are in clinical trial. To fasten the drug approval procedure and to discover more efficacious inhibitors with novel scaffolds that can improve the anticancer therapeutics status, computational drug discovery approaches are highly reliable. Current study aimed at phytochemical screening for potential CDK2 inhibitor. Idea of using phytochemicals instead of synthetic compounds for drug screening is to minimize the multifaceted side effects posed by multitargeting of other signalling proteins in addition to CDK2. Medicinal plants have been used for centuries to cure different diseases [33-35]. Medicinal plants produce different secondary metabolites generally called phytochemicals (flavonoids, alkaloids, terpenoids, polyphenolics etc) which have anticancer potential and can be used as an alternative drug resource [15,32].
Form the ground-breaking detail of structural diversity among natural products, virtual screening was done to discover novel allosteric compound for regulation of dysfunctional CDK2. Allosteric regulation has been reported as effective strategy to attain irreversible inhibition. Studies reports that high similarity within the ATP binding site of CDK2 with other cell cycle signalling protein results in off target binding of inhibitors [29,36]. To deal with the daunting task of selectivity and specificity of CDK2 inhibitors, current study is devoted to allosteric regulation for desired results.
Spatial orientation and dock score of current reported four top-ranked leads reveals efficient binding of functional residues with maximum binding affinity. Moreover, these compounds meet the drug likelihood criteria shown in Table 1 and may prove excellent ready to use starting point. A study by Lima and co-workers established the role Khusenol T as anti-inflammatory and antinociceptive which is thought to relief pain in pathological disease [37]. Antimicrobial potential of Remangiflavanone B has also been reported in past study [24], while Neocalyxins A is known for its cytotoxic role in tumor cells [25]. In addition to this Elenoside has been reported as a cytotoxic bioactive molecule in in vitro. Along with this central nervous activity and its role as cardiac muscles vasodilator has also been reported [26]. Looking at the anticancer drug potential of aforementioned phytochemicals, current study was an endeavour to exploit the chemical nature of four selected phytochemicals for inhibitions of limitless proliferation of tumour cells by inhibiting CDK2 protein.
Current molecular docking study has shown the important interactions of medicinal plant phytochemicals with CDK2 binding site. Kushenol T, Remangiflavanone B, Neocalyxins A and Elenoside showed the low S score as compared to the ANS and also interacts strongly with all eight residues of CDK2 binding site. These phytochemicals successfully block the CDK2. Thus, it can be concluded from this study that these phytochemicals can be used as strong anti-cancer agents against CDK2 in future.
Conflict of Interest
The authors declare no conflict of interest.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell, (2011); 144(5): 646-674.
- Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Proliferation, (2003); 36(3): 131-149.
- Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene, (2009); 28(33): 2925.
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nature Reviews Cancer, (2009); 9(3): 153.
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nature Reviews Drug Discovery, (2009); 8(7): 547-566.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians, (2016); 66(1): 7-30.
- McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clinics in Liver Disease, (2015); 19(2): 223-238.
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, et al. Cancer treatment and survivorship statistics, 2016. CA: A Cancer Journal for Clinicians, (2016); 66(4): 271-289.
- Vecchia C, Malvezzi M, Bosetti C, Garavello W, Bertuccio P, et al. Thyroid cancer mortality and incidence: a global overview. International Journal of Cancer, (2015); 136(9): 2187-2195.
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nature Reviews Drug Discovery, (2015); 14(2): 130.
- Varbanov HP, Kuttler F, Banfi D, Turcatti G, Dyson PJ. Repositioning approved drugs for the treatment of problematic cancers using a screening approach. PloS One, (2017); 12(2): e0171052.
- Kinch MS. An analysis of FDA-approved drugs for oncology. Drug Discovery Today, (2014); 19(12): 1831-1835.
- Wu P, Nielsen TE, Clausen MH. Small-molecule kinase inhibitors: an analysis of FDA-approved drugs. Drug Discovery Today, (2016); 21(1): 5-10.
- Sherr CJ, Bartek J. Cell Cycle–Targeted Cancer Therapies, (2017); 1: 41-57.
- Shukla S, Mehta A. Anticancer potential of medicinal plants and their phytochemicals: a review. Brazilian Journal of Botany, (2015); 38(2): 199-210.
- Avni GD, Ghulam NQ, Ramesh KG, Mahmoud E-T, Jaswant S, et al. Medicinal Plants and Cancer Chemoprevention. Current Drug Metabolism, (2008); 9(7): 581-591.
- David MP, Patricia V, Georgina C-d-S, Natercia T, Paula BA. Plant Secondary Metabolites in Cancer Chemotherapy: Where are We? Current Pharmaceutical Biotechnology, (2012); 13(5): 632-650.
- Chemical Computing Group ULC (2013) Molecular Operating Environment (MOE) 09. 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2017.
- Mumtaz A, Ashfaq UA, ul Qamar MT, Anwar F, Gulzar F, et al. MPD3: a useful medicinal plants database for drug designing. Natural Product Research, (2017); 31(11): 1228-1236.
- Ashfaq UA, Mumtaz A, Qamar Tu, Fatima T. MAPS Database: Medicinal plant Activities, Phytochemical and Structural Database. Bioinformation, (2013); 9(19): 993-995.
- Irwin JJ, Shoichet BK. ZINC− a free database of commercially available compounds for virtual screening. Journal of Chemical Information and Modeling, (2005); 45(1): 177-182.
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annual Reports in Computational Chemistry, (2008); 4217-241.
- Betzi S, Alam R, Martin M, Lubbers DJ, Han H, et al. Discovery of a Potential Allosteric Ligand Binding Site in CDK2. ACS Chemical Biology, (2011); 6(5): 492-501.
- Deng Y, Lee JP, Tianasoa-Ramamonjy M, Snyder JK, Des Etages SA, et al. New Antimicrobial Flavanones from Physena madagascariensis. Journal of Natural Products, (2000); 63(8): 1082-1089.
- Minh Giang P, Tong Son P, Matsunami K, Otsuka H New Diarylheptanoids from Alpinia pinnanensis. Chemical and Pharmaceutical Bulletin, (2005); 53(10): 1335-1337.
- Navarro E, Alonso SJ, Trujillo J, Jorge E, Pérez C. Central nervous activity of elenoside. Phytomedicine, (2004); 11(6): 498-503.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1PII of original article: S0169-409X(96)00423-1. The article was originally published in Advanced Drug Delivery Reviews 23 (1997) 3–25.1. Advanced Drug Delivery Reviews, (2001); 46(1): 3-26.
- Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell, 100(1): 57-70.
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature, (2009); 458(7239): 719.
- Ahmed B, Ashfaq UA, ul Qamar MT, Ahmad M. Anticancer potential of phytochemicals against breast cancer: Molecular docking and simulation approach. Bangladesh Journal of Pharmacology, (2014); 9(4): 545-550.
- O'leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nature Reviews Clinical Oncology, (2016); 13(7): 417-430.
- Pezzuto JM. Plant-derived anticancer agents. Biochemical Pharmacology, (1997); 53(2): 121-133.
- ul Qamar MT, Mumtaz A, Ashfaq UA, Adeel MM, Fatima T. Potential of plant alkaloids as dengue ns3 protease inhibitors: Molecular docking and simulation approach. Bangladesh Journal of Pharmacology, (2014); 9(3): 262-267.
- ul Qamar MT, Mumtaz A, Rabbia Naseem AA, Fatima T, Jabbar T, et al. Molecular docking based screening of plant flavonoids as dengue NS1 inhibitors. Bioinformation, (2014); 10(7): 460.
- ul Qamar T, Mumtaz A, Ashfaq UA, Azhar S, Fatima T, et al. Computer aided screening of phytochemicals from garcinia against the dengue NS2B/NS3 protease. Bioinformation, (2014); 10(3): 115.
- Wu P, Nielsen TE, Clausen MH. FDA-approved small-molecule kinase inhibitors. Trends in Pharmacological Sciences, (2015); 36(7): 422-439.
- Lima GM, Quintans-Júnior LJ, Thomazzi SM, Almeida EMSA, Melo MS, et al. Phytochemical screening, antinociceptive and anti-inflammatory activities of Chrysopogon zizanioides essential oil. Revista Brasileira de Farmacognosia, (2012); 22443-450.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0/