Research Article
Occurrence of HCV genotypes in different age groups of patients from Lahore, Pakistan
Amna Rasheed1, Sajjad Ullah1, Sajid Naeem2*, Muhammad Zubair3, Waseem Ahmad3, Zahid Hussain2
Adv. life sci., vol. 1, no. 2, pp. 89-95, February 2014
*-Corresponding Author: Sajid Naeem (meharsajidnaeem@hotmail.com)
Author Affiliations
2-Institute of Industrial Biotechnology Govt. College University, Lahore, Lahore – Pakistan
3-Institute of Molecular Biology and Biotechnology, University of Lahore, Lahore – Pakistan
Abstract
Introduction
Methods
Results
Discussion
References
Abstract
Background: Hepatitis C virus is a small, enveloped single stranded, positive sense RNA virus. Different genotypes are distributed in different geographical areas of the world. Determination of HCV genotype is a powerful tool for the treatment of chronic and acute liver disease.
Method: The present study was carried out to find the occurrence of different HCV genotypes in the city of Lahore, a populous city of Pakistan from January 2010 to December 2010. Blood sample of patients positive for anti HCV by ELISA as well as HCV by PCR were collected and plasma was separated. HCV viral RNA load was analyzed in these samples using Real Time PCR. Qiagen HCV mini kit for RNA extraction and Qiagen HCV amplification kit for PCR amplification were used. Amplicons were subjected to HCV genotyping using Third Wave Technology.
Results: Among 489 patients, 211 (43.1%) patients were males and 278 (56.9%) were females. Occurrence of HCV in the age group of 36-45 years was 32.5 %. Occurrence of HCV genotype 1 was 9.6% (47), genotype 3a was 80.77% (395), genotype 3h was 1.0% (5) , genotype 4 was 4.9% (24), co-infection of genotypes 1 & 2 was 0.2% (01), co-infection of genotypes 1 & 3 was 0.6% (03) and co-infection genotypes 1 & 4 was 0.4% (02).
Conclusion: HCV genotype 3a is most prevalent HCV genotype in subjected population during said duration with most infected people from 26 to 35 years of age. Female population is having more of HCV infection as compared to males.
Keywords: Hepatitis C virus, Lahore, Occurrence, Genotype 3a, ELISA, PCR
Introduction
Infection with Hepatitis C Virus (HCV) is estimated at more than 200 million people worldwide, representing more than 3% of the world's population [1]. HCV consists of an envelope derived from host membranes into which are inserted the virally encoded glycoproteins E1 and E2, surrounding a nucleocapsid and a positive sense single stranded RNA genome which has been characterized and sequenced [2]. Phylogenetic analysis of HCV sequence has revealed 6 major genotypes and more than 50 subtypes [3,4]. HCV genotypes differ from each other in their nucleotide sequence by 31-34% and in their amino acid sequence by 30% [5]. The genotype determination is a relevant clinical practice. It helps to predict the probability of sustained virological response i.e. 40%-45% for genotype 1 as compared with 70%-80% for genotypes 2 and 3. It is also used routinely to determine duration of treatment i.e. 48 weeks for genotypes 1 and 4, and 24 weeks for genotypes 2 and 3 [6].
Genotypes 1, 2 and 3 are distributed almost worldwide [7,8]. Half of the hepatitis C patients from South of Brazil were infected by genotypes 2 and 3 [9]. Types 4, 5 and 6 have been found in distinct geographical areas [3,8,10]. Genotype 5 is mainly dominant in South Africa and also present in less percentage in other countries [11]. Genotype 6 is common in South East Asia, Asian countries and Asian Australians [12]. In Romania, HCV subtype 1a was 5.4 %, subtype 1b was 92.6%, subtype 3a was 0.8% and subtype 4a was 1.2% [13]. In the Swat district of Pakistan, HCV genotype 3a had been reported to be predominant followed by mixed genotype infection and then 3b [14]. Genotype 7, 8 and 9 had been reported from Vietnam [15]. Genotype 10 and 11 were identified in patients from Indonesia but there had been a conflict for the classification of HCV isolate and scientists proposed that genotypes 7 through 11 must be considered as variants of the same group and must be classified as a single genotype, type 6 [15,16]. From Pakistan, few studies are available on the distribution of various HCV genotypes and its route of transmission based on small sample sizes [17-19].
Methods
All the work was done in PCR section of Pathology Department, Jinnah Hospital, Lahore from January 2010 to December 2010. Three milliliter blood sample was collected from each of 489 HCV positive patients along with their age, gender and place of living. Plasma was separated from each sample using the recommended standard procedure given by Qiagen extraction kit. Extraction of HCV RNA from each sample was done using QIAamp Viral mini kit by Qiagen cat no. 52906. Amplification was achieved by using Atrus HCV RT-PCR kit by Qiagen from Germany. HCV genotyping of amplified samples was detected using Invader HCV Reagents on Palm Cycler for amplification and Cytoflour for detection of genotype. The Invader® chemistry is composed of two simultaneous isothermal reactions. A primary reaction specifically and accurately detects single-base changes, insertions, deletions and changes in gene and chromosome number for genetic, pharmacogenetic and infectious diseases. A second reaction is used for signal amplification and generic Present study was carried out to find the occurrence of different HCV Genotypes in the city of Lahore, Punjab of Pakistan. In the present study a total number of 489 HCV positive cases (which were also anti HCV positive by ELISA) were selected. HCV load was analyzed in these samples using Real Time PCR. Genotypes of all samples were determined using Third Wave Technology.
Results
Among 489 patients 211 (43.1%) patients were male and 278 (56.9%) were female. Out of 211 (43.1%) anti HCV positive males, 24 (4.9%) had HCV genotype 1, 168 (35.4%) had HCV genotype 3a, 03 (0.6%) had HCV genotype 3h, 10 (2.0%) had HCV genotype 4, 01 (0.2%) had co-infection of genotypes 1 & 3 and un-typable were 5 (1.02%). Among 278 (56.9%) anti HCV positive females, 23 (4.7%) had HCV genotype 1, 227 (46.4%) had HCV genotype 3a, 02 (0.4%) had HCV genotype 3h, 14(2.9%) had HCV genotype 4, 01 (0.2%) had co-infection of genotypes 1 & 2, 02 (0.4%) had co-infection of genotypes 1 & 3 and 02 (0.4%) had co infection of genotypes 1 & 4 and untypable were only 7 (1.43%) (Table I).
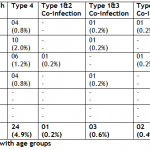
Out of 489 patients, 15.5% (76) were from 15-25 years age group. Out of which 2.0% (10) had HCV genotype 1, 12.3% (60) had HCV genotype 3a, 0.8% (04) had HCV genotype 4, 0.2% (01) was co infected with HCV genotypes 1&3 and 0.2% (01) was co infected with HCV genotypes 1 & 4. Out of 489 patients, 33.9% (166) were from 26-35 years age group. Out of which 2.9% (14) had HCV genotype 1, 27.4% (134) had HCV genotype 3a, 0.6% (03) patients had HCV genotype 3h, 2.0% (10) had HCV genotype 4 and 0.2% (01) was co infected with genotypes 1 & 3. Out of 489 patients, 32.5% (159) were from 36-45 years age group. Out of which 2.5% (12) had HCV genotype 1, 27.4% (134) had HCV genotype 3a, 0.2% (01) had HCV genotype 3h, 1.2% (06) had HCV genotype 4, 0.2% (01) was co infected with HCV genotypes 1 & 2 and 0.2% (01) was co-infected with HCV genotypes 1 & 4. Out of 489 patients, 13.5% (66) were from 46-55 years age group, out of which 1.7% (08) had HCV genotype 1, 10.6% (52) had HCV genotype 3a, 0.2% (01) had HCV genotype 3h, 0.8% (04) had HCV genotype 4 and 0.2% (01) was co-infected with HCV genotypes 1 & 3. Out of 489 patients, 3.7% (18) were from 56-65 years age group. Out of which only 0.4% (02) had HCV genotype 1 and 3.3% (16) had HCV genotype 3a. 489 0.8% (04) patients were from > 65 years age group. Out of which 0.2% (01) had HCV genotype 1 and 0.6% (03) had HCV genotype 3a (Table II).
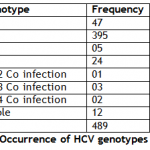
Among 489 anti HCV positive cases on ELISA, occurrence of HCV genotype 1 was 9.6% (47) , genotype 3a was 80.77% (395) , genotype 3h was 1% (05) , genotype 4 was 4.9% (240) , co infection of genotypes 1 & 2 was 0.2% (01) , co infection of genotypes 1 & 3 was 0.6% (03) and co infection genotypes 1 & 4 was 0.4% (02) and un-typable were 2.45% (12) (Table III).
Discussion
Interest of HCV genotyping by group screening is increased many fold as it is useful for the solution of epidemiological questions and development of vaccines against HCV. It has also been shown to be valuable to facilitate therapeutic decisions and strategies [20,21]. Severity, progression and response to the treatment of disease caused by HCV vary according to the genotype of HCV [22,23].
Knowledge of occurrence of different HCV genotypes in different areas of Pakistan will help in theraputic implications. In the present study, occurrence of various genotypes of HCV, in Lahore city, were observed. All plasma samples were anti HCV positive on ELISA and thus were further tested for HCV genotyping. The data shows that HCV infection is more common in females residing in Lahore as compared to males [20]. Results were analyzed by keeping the age and gender categorical variable and patients were divided in six different age groups. It is found that patients belonging to age group 26-35 years are more infected with HCV followed by the age group of 36-45 years. A similar reported same kind of results that people ≤ 40 years of age were more affected with HCV in comparison to those > 40 years of age in Lahore [24]. A study reported that HCV prevalence observed was highest among an age group of 13-50 years [25]. In present study prevalence of HCV genotypes is also recorded with reference to gender and it is found that highest prevalence of type 3 exists among both genders. Genotype 3a predominates among the patients residing in Lahore, genotype 1 and genotype 4 follows it. This data favors the results of Ahmad who reported that the most commonly detected genotype in their study was genotype 3 (59.1%), with predominant subtype 3a (55.9%) and 3b (3.2%) [24]. Genotype 1a was (23.6%) while genotype 4 (13.7%) comprised of the subtypes a (12.5%) and b (1.2%). It is also reported the occurrence of HCV genotype 3a in 40.96%, 3b in 15.66%, 1a in 9.63%, and 1b in 2.40% of hepatocellular carcinoma tissue (HCT) samples [26]. They also found 24 (28.91%) mixed types. Another study by Butt et al., in 2010 showed prevalence of 3a (62%), 3b (9%), 1a (3%), 2a (2.144%), mixed (4.718%) and un type able (17.16%) [27]. Another study reported that genotype 1 was the most frequently genotype found in all regions of Brazil. In present study prevalence of mixed infection is quite negligible [28].
This is dire need to carry out a detailed study on patterns of HCV genotype amongst the population of Lahore for the proper and better control on spread of disease.
Conclusively, anti HCV positivity was found relatively more predominant in females than male population. HCV RNA was common in the age group of 26-35 followed by the age group of 36-45 years. Genotype 3a was the common genotype found in the population of the city of Lahore, Pakistan and it was also the common genotype in both males and females.
Competing interests
All the authors have no financial or non-financial competing interests.
Data and Tables
Table I: Distribution of HCV genotypes with genders Table II: Distribution of HCV genotypes with age groups Table III: Occurrence of HCV genotypes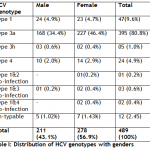
- Pavio N, Lai MM. The hepatitis C virus persistence: how to evade the immune system? Journal of biosciences, (2003); 28(3): 287-304.
- Bhandari BN, Wright TL. Hepatitis C: an overview. Annual review of medicine, (1994); 46309-317.
- Yap P, Kolberg J, Urdea M. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. Journal of General Virology, (1993); 742391-2399.
- Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology, (2005); 42(4): 962-973.
- Pawlotsky J-M. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antiviral research, (2003); 59(1): 1-11.
- Saracco G, Ciancio A, Olivero A, Smedile A, Roffi L, et al. A randomized 4‐arm multicenter study of interferon alfa–2b plus ribavirin in the treatment of patients with chronic hepatitis C not responding to interferon alone. Hepatology, (2001); 34(1): 133-138.
- McOmish F, Yap P, Dow B, Follett E, Seed C, et al. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. Journal of Clinical Microbiology, (1994); 32(4): 884-892.
- Chan S, McOmish F, Holmes E, Dow B, Peutherer J, et al. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. The Journal of general virology, (1992); 731131-1141.
- Silva CMDd, Costi C, Krug LP, Ramos AB, Grandi T, et al. High proportion of hepatitis C virus genotypes 1 and 3 in a large cohort of patients from Southern Brazil. Memórias do Instituto Oswaldo Cruz, (2007); 102(7): 867-870.
- Bukh J, Purcell RH, Miller RH. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proceedings of the National Academy of Sciences, (1993); 90(17): 8234-8238.
- Bonny C, Fontaine H, Poynard T, Hézode C, Larrey D, et al. Effectiveness of interferon plus ribavirin combination in the treatment of naive patients with hepatitis C virus type 5. A French multicentre retrospective study. Alimentary pharmacology & therapeutics, (2006); 24(4): 593-600.
- Pybus OG, Markov PV, Wu A, Tatem AJ. Investigating the endemic transmission of the hepatitis C virus. International journal for parasitology, (2007); 37(8): 839-849.
- Sultanat C, Oprisan G, Szmal C, Vagu C, Temereanca A, et al. Molecular epidemiology of hepatitis C virus strains from Romania. Journal of Gastrointestinal & Liver Diseases, (2011); 20(3).
- Ali A, Ahmed H, Idrees M. Molecular epidemiology of Hepatitis C virus genotypes in Khyber Pakhtoonkhaw of Pakistan. Virology Journal, (2010); 7(1): 203.
- Tokita H, Okamoto H, Tsuda F, Song P, Nakata S, et al. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proceedings of the National Academy of Sciences, (1994); 91(23): 11022-11026.
- de Lamballerie X, Charrel RN, Attoui H, De Micco P. Classification of hepatitis C virus variants in six major types based on analysis of the envelope 1 and nonstructural 5B genome regions and complete polyprotein sequences. Journal of General Virology, (1997); 78(1): 45-51.
- Abdulkarim AS, Zein NN, Germer JJ, Kolbert CP, Kabbani L, et al. Hepatitis C virus genotypes and hepatitis G virus in hemodialysis patients from Syria: identification of two novel hepatitis C virus subtypes. The American journal of tropical medicine and hygiene, (1998); 59(4): 571-576.
- Idrees M. Detection of Six Serotypes of HCV in anti-HCV Positive Patients and rate of ALT/AST abnormalities. Pak J Microbiol, (2001); 261-65.
- Idrees M, Riazuddin S. Frequency distribution of hepatitis C virus genotypes in different geographical regions of Pakistan and their possible routes of transmission. BMC Infectious Diseases, (2008); 8(1): 69.
- Poynard T, Bedossa P, Chevallier M, Mathurin P, Lemonnier C, et al. A comparison of three interferon alfa-2b regimens for the long-term treatment of chronic non-A, non-B hepatitis. New England Journal of Medicine, (1995); 332(22): 1457-1463.
- McHutchison J, Poynard T, Davis G, Esteban-Mur R, Harvey J, et al. Evaluation of hepatic HCV RNA before an after treatment with interferon alfa 2B or combined with ribavirin in chronic hepatitis; 1999. WB SAUNDERS CO INDEPENDENCE SQUARE WEST CURTIS CENTER, STE 300, PHILADELPHIA, PA 19106-3399 USA. pp. 363A-363A.
- Dusheiko G, Main J, Thomas H, Reichard O, Lee C, et al. Ribavirin treatment for patients with chronic hepatitis C: results of a placebo-controlled study. Journal of hepatology, (1996); 25(5): 591-598.
- Nousbaum J-B, Pol S, Nalpas B, Landais P, Berthelot P, et al. Hepatitis C virus type 1b (II) infection in France and Italy. Annals of internal medicine, (1995); 122(3): 161-168.
- Ahmad W, Ijaz B, Javed FT, Jahan S, Shahid I, et al. HCV genotype distribution and possible transmission risks in Lahore, Pakistan. World journal of gastroenterology: WJG, (2010); 16(34): 4321.
- Ali M, Kanwal L, Tassaduqe K, Iqbal R. Prevalence of Hepatitis C Virus (HCV) in Relation to Its Promotive Factors Among Human Urban Population of Multan, Pakistan. European Journal of General Medicine, (2009); 6(2).
- Idrees M, Riazuddin S. A study of best positive predictors for sustained virologic response to interferon alpha plus ribavirin therapy in naive chronic hepatitis C patients. BMC gastroenterology, (2009); 9(1): 5.
- Butt S, Idrees M, Akbar H, Awan Z, Afzal S, et al. The changing epidemiology pattern and frequency distribution of hepatitis C virus in Pakistan. Infection, Genetics and Evolution, (2010); 10(5): 595-600.
- Campiotto S, Pinho J, Carrilho F, Da Silva L, Souto F, et al. Geographic distribution of hepatitis C virus genotypes in Brazil. Brazilian Journal of Medical and Biological Research, (2005); 38(1): 41-49.




