Review Article
Human Cryptosporidiosis: An insight into Epidemiology, Modern Diagnostic Tools and Recent Drug Discoveries
Arsalan Zafar1, Muhammad Kasib Khan1*, Zaheer Abbas1, Rao Zahid Abbas1, Zia ud Din Sindhu1, Zafar Iqbal1, Muhammad Shafi Hasni1, Hasnain Javed2, Muhammad Nadeem1, Hammad Ur Rehman Bajwa1, Muhammad Adnan Sabir Mughal1, Fatima Yasin1
Adv. life sci., vol. 6, no. 2, pp. 60-70, February 2019
*- Corresponding Author: Muhammad Kasib Khan (Email: mkkhan@uaf.edu.pk)
Authors' Affiliations
2- Punjab AIDS Control Program, Health Department, Government of Punjab, Pakistan
Abstract![]()
Introduction
Methods
Discussion
Conclusion
References
Abstract
Cryptosporidiosis is an emerging food and water borne zoonotic disease, which is caused by genus Cryptosporidium. The first Cryptosporidium spp. was isolated from mice in 1907 and gained importance when it was found in an HIV positive patient. It usually causes self-limiting diarrhea in young children and immunocompetent patients. However, it may lead to chronic diarrhea with life threatening condition in immunocompromised patients. Other complications related to this transmittable infection may include respiratory problems, skin rashes and headache. HIV/AIDS patients are highly susceptible host for this parasite. Cryptosporidium parvum and Cryptosporidium hominis are the known pathogenic species, prevalent among humans and they are being transmitted through contaminated food and water. Usually, the diagnosis of Cryptosporidium spp. is dependent on microscopic technique in many countries, which has a low sensitivity and specificity leading to false positive results. However, for a step forward to successful epidemiological studies, advanced techniques (Serological and DNA-based) provide us the better ways of diagnosis with more sensitivity and specificity. Furthermore, no antiparasitic drug has found to be effective against Cryptosporidium spp. except Nitazoxanide which is FDA-approved and effective only when administered along with antiretroviral therapy. In this regard, present review summarizes the various epidemiological studies conducted around the globe along with modern diagnostic tools and the suitable treatment available now a days. This systemized review will help the scientists to better understand all the aspects of cryptosporidiosis at one platform which may help in designing surveillance studies through selection of sensitive diagnostic techniques. The new drugs mentioned in this review may also help to better control this parasite in humans, especially immunocompromised individuals.
Keywords: Cryptosporidium; Opportunistic protozoa; Zoonosis; Recent advancements
Cryptosporidium spp., an opportunistic intracellular protozoan parasite, belong to Phylum Apicomplexa and causes high economic losses in case of morbidity and mortality by affecting the gastrointestinal tract of sheep, goat, pig, cattle and human [1]. The first specie was isolated from gastric juice of mice, named as Cryptosporidium muris, in 1907 by Tyzzer [2]. Later, Cryptosporidium was also detected, from human stool sample in 1976 and became most emergent protozoa due to its zoonotic importance. Despite, Cryptosporidium spp. are host specific, there are 26 recognized species causing chronic diarrhea, dysentery and malabsorption in humans as well as in farm animals. Most prevalent species in humans are C. parvum and C. hominis. However, new species or new strains are going to be discovered due to rapid genetic recombination as i) two or more species are affecting the same host at the same time ii) it undergoes both sexual and asexual production leading to genetic variation [3].
Chances of infection; chronic diarrhea/dysentery, are more in immunocompromised patients like HIV patients as compared to Immunocompetent patients; who suffer with a short-term and self-limiting diarrhea [4,5]. In the past, Cryptosporidium was considered to be linked with childhood diarrhea, malnutrition. Situation became worse in 1993 when 4 million people got Cryptosporidium infection in USA and Milwaukee. Despite of the evidence, cryptosporidiosis was under-diagnosed, and the treatment was only symptomatic [6].
Nevertheless, new diagnostic tools and literature has improved the estimate of global burden. However, there are still gaps which need to be filled, particularly in the fields of epidemiological studies, diagnosis and proper cure for cryptosporidiosis. Moreover, there is a deficiency of well-organized and comprehensive reviews covering all the aspects of cryptosporidiosis i.e. disease outbreaks, modern diagnostic tools and therapeutic controls especially in HIV patients. Such detailed reviews are very important for the young parasitologists and researchers for better understanding the current scenarios regarding the disease epidemics, modern diagnostic tools and the latest drug discoveries. Considering all the above-mentioned prospects, following review was planned to summarize a detailed information about the disease burden of Cryptosporidium infection in many countries, modern diagnostic tools and the new drugs available against Cryptosporidiosis.
Literature survey and selection criteria
The main objective of writing this review was to summarize recent advancements in some aspects of cryptosporidiosis important for controlling the disease, including epidemiology and recent disease outbreaks in various countries, virulent factors, latest diagnostic techniques and recently available drugs to cure Cryptosporidium infection, particularly in HIV/AIDS patients. Doing so, a massive search was carried out on google scholar by providing different terms like “Cryptosporidiosis”, “Disease outbreaks of Cryptosporidium parvum”, “Epidemiological studies on Cryptosporidium parvum”, “Cryptosporidium infection is HIV/AIDS patients”, “Cryptosporidium infection in children”, “diagnostic techniques in Cryptosporidium infection”, “modern techniques for the identification of Cryptosporidium parvum”, “DNA based techniques for the identification of Cryptosporidium in humans”, “Mortality rate in immunocompromised patients due to Cryptosporidium spp.”, “how to treat Cryptosporidium infection in humans”, “latest drug discoveries against Cryptosporidium infection”, “FDA approved drugs for cryptosporidiosis”. A total of 65 citations were reviewed after screening according to the contents of this review.
Epidemiology
Early epidemiological studies have revealed the prevalence of Cryptosporidium to be 1% in developed countries while 5-10% in poor countries [6]. However, results of recent studies, based on serological and molecular techniques, revealed that the previous studies have underestimated the frequency of C. parvum infection indicating the prevalence rate of 15-25% in diarrheal patients [6].
Disease burden
Numerous researches have been conducted on Cryptosporidiosis throughout the world due to its zoonotic and opportunistic behavior. Regarding the global disease burden published in 2010, highest prevalence rate of 3.5-35.8% has been documented in America, followed in order by Africa (2.6-21.3%), Europe (0.1-14.4%) and Asia (1.3-13.1%) [7]. However, a brief history of current occurrence of cryptosporidiosis documented in various hosts along with various diagnostic techniques is listed in Table 1.
Transmission and Pathogenesis
C. parvum mostly transmit either by direct or indirect ways. Direct transmission occurs when humans drink contaminated water or food with infected fecal material or by direct touching the infected person. Chances of disease spread are higher in day care centers and schools, where children get exposed to the contaminated surface and mud while playing. Meanwhile, a person can also get infection with sexual practices. Moreover, indirect transmission may also occur by drinking or eating contaminated water and food, respectively. It is clear with evidence that C. parvum can easily be passed through water filtrations plants due to its very small size of one micron. Furthermore, there is no effect of chlorination on these parasites during water treatment. So, drinking water is a main source of its transmission [13,41,42].
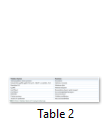
Pathologically, Cryptosporidium cause inflammatory damage to the epithelium of small intestine, which leads to the malabsorption and increased intestinal secretions resulting in diarrhea, dysentery, vomiting and nausea which may last for more than seven days. Situation may lead to death due to sever and persistent diarrhea in very young children and immunocompromised/HIV patients. By entering the host, they attach with intestinal epithelium with the help of gp900 and gp60 (glycoproteins) and start proliferating with the help of micronemes and rhoptries, resulting in the formation of parasitophorous vacuoles where they undergo sexual and asexual reproduction. The whole situation is called cryptosporidiosis [42]. Some studies have shown that Cryptosporidium may also affect the respiratory tract and cause some respiratory disorders along with skin rashes and headache [4,7,43]. Several virulenec factors cause damage to the host cells are mentioned in Table 2 [1].
Associated risk factors
It is evident from various studies that many associated risk factors may likely to be linked with occurrence of cryptosporidiosis in humans. Some of them found associated with varied prevalence of cryptosporidiosis have been listed below:
Seasonal variations
Cryptosporidiosis has been found endemic throughout the year in various parts of the world, but infection is most likely to be happened in rainy season due to water contamination [44]. In a critical study, another aspect was highlighted when Cryptosporidium along with Giardia and Entamoeba showed clear seasonal variation. The prevalence was stable before rainy season but during the rainy season, an increased rate of infection was noticed. However, this analysis does not support the pattern in case of cattle host.
Age
As a result of cryptosporidiosis, different immune responses can be observed which are dependent on age variation of host. Cryptosporidiosis is found to be more prevalent in children of age less than 24 months as they have less developed immune system as compared to adults [11]. Some supporting results were obtained in a research conducted at a goat farm in Brazil. This study showed more prevalence of Cryptosporidium in juvenile (age less than 12 months) as compared to adults (age more than 12 months) as all 4.8% prevalence was observed in juvenile animals [45]. To observe the prevalence of cryptosporidiosis, a farm level study was performed which revealed the risk to be high in calves of age 8-21 weeks in comparison with 0-7 days aged calves. During the same year, similar results were found in buffalo calves which showed high risk of prevalence of infection in calves of age 1-15 days [46]. In another farm level study, PCR study disclosed cryptosporidiosis to be more prevalent in pre-weaned calves as compared to post weaned calves. In human, Cryptosporidium was comparatively more prevalent in children of age 7-15 days than children of 3-12 years.
Gender
Occurrence of cryptosporidiosis is higher in male population as compared to female population. However, it may vary due to the exposure level of males and females. In china, It has been observed that the prevalence of Cryptosporidium infection was 3 times higher in males (12.6%) as compared to those of females (4.4%) [47].
Socio-economics behavior
Socio-economic behavior of host directly affects the transmission of Cryptosporidium. Cryptosporidium is transmitted through ingestion of parasites which could be by means of waterborne, person to person and food borne whereas the middle one is occasional. The cryptosporidium mostly affects children with immunosuppression while suppression is more prominent in malnourished children as compared to children aged less than 6 months which are breastfed [48]. In a study, it was observed that cryptosporidium spp., were more prevalent in rural areas especially in the persons exposed to public toilets and infected with HIV [49]. In another investigation, samples from 1731 drug abuser rehabilitation patients were collected to record prevalence of Cryptosporidium spp., infection and the results showed 19.6% persons to be positive with infection.
Diagnostic Tools
- Conventional Method
Traditionally, Cryptosporidium oocysts detection in environment, food, water, tissue and fecal sample has greatly relied on microscopic examination. But when it comes to the identification of Cryptosporidium, morphological characters are few. Therefore, identification based on light microscopy is time consuming and unreliable. Several staining techniques are used to identify Cryptosporidium oocysts. The least simple stains include modified Ziehl–Neelsen, Kinyoun, dimethyl sulphoxide carbol fuchsin and safranin–methylene blue. However, samples containing less oocysts number can suffer low sensitivity and/or specificity. Moreover, negative staining methods are also available. For example, by using chemicals like malachite green and green perbromide, background of slide is stained leaving the oocysts unstained. These latter staining techniques are time consuming, laborious and require perfection. Moreover, none of the techniques is helpful to identify Cryptosporidium species. For the identification of Cryptosporidium oocysts, modified Ziehl-Neelsen staining and wet mount preparation methods are also commonly used [36]. Another method for the identification and detection of Cryptosporidium oocysts is flow cytometry. To identify Cryptosporidium in sewage samples and water, Vesey used a skytron Argos 100-5 instrument spiked with oocysts stained with an FITC-C-mAb. The outcome of this study is encouraging by means of flow cytometry because it detects as few as 1000 oocysts/L.
- Immunological Assays
For detection and identification, Immunological methods have several advantages over microscopic techniques. For instance, direct fluorescent antibody (DFA) and fluorescence microscopy assay use fluorescence in isothiocyanate-conjugated anti-Cryptosporidium monoclonal antibody (FITC-C-mAb), which identify epitopes on the surface of Cryptosporidium oocysts with high sensitivity (98.5–100%) and specificity (96–100%) for Cryptosporidium oocysts detection in environmental samples and fecal smears. FITC-C, commercially available monoclonal antibody, is used for enumerating and detecting Cryptosporidium oocysts routinely in environmental and fecal samples. DFA assays which are based on monoclonal antibody can differ in their sensitivity and specificity of diagnosis which depends on many factors including ‘biophysics’ detection system, antibody conjugated reporter (enzyme fluorochrome), Cryptosporidium antigen used to develop mAb and the avidity and subclass/class of the antibody. IgG monoclonal antibody seems to have better avidity to Cryptosporidium oocyst surface antigens as compared to IgM resulting in better recoveries with immuno-magnetic separation (IMS) within water samples of high turbidity. Both molecular interaction and fluorochrome selection are the critical aspects and are responsible for false positive results due to autofluorescence in emission and excitation spectra. Though different monoclonal antibodies have been developed but commercially existing mAbs cannot identify Cryptosporidium specifically. Monoclonal antibodies have been developed with lesser number of Cryptosporidium parvum oocysts [50].
In the last few years, identification and diagnostic trends have been greatly changed towards EIA (Enzyme immunoassays), IC (Immuno-chromography) and ELISA (Enzyme Linked Immunosorbent Assay) for the detection of Cryptosporidium oocyst antigens. Specificity of these assays is reported to be high (98–100%). Advantage of these assays is that they detect infections when oocysts are not being excreted in feces [51]. while limitation of some kits may be seen because they mainly depend upon visual inspection which enable subjective interpretation of test results.
Furthermore, these assays like many other immunological approaches do not diagnose Cryptosporidium genotype or species. So far, no such immunological tool is present which makes a distinction among oocysts of dissimilar species. One effort has been made with the development of PCR-EIA which differentiates between C. hominis and C. parvum. In a recent study, results have shown the 100% specificity and 86.6% sensitivity of antigen ELISA for Cryptosporidium [52].
- DNA Based Approaches
Molecular techniques are available to find out species, genotypes and sub-genotypes of Cryptosporidium to differentiate among human and animal pathogens. Nucleic acid-based methods have been evaluated for species identification in animals and human from environmental, water and fecal samples. Some methods are based on in-situ hybridization of probes within oocysts to specific genetic loci, while most methods are based on specific amplification of loci from genomic DNA by PCR. These applications have led to understanding Cryptosporidium species regarding biology, ecology, epidemiology, systematic and population genetics for control and prevention of cryptosporidiosis in animals and humans. DNA profiles from human isolates showed two different genotypes, C. hominis (anthroponotic) and C. parvum zoonotic genotype. Analysis of tri-nucleotide sequence has shown twelve subtypes of Cryptosporidium hominis and seven subtypes of Cryptosporidium parvum [53].
FISH (Fluorescent In-situ Hybridization) utilizes probes (fluorescently-labelled oligonucleotides) by targeting RNA or DNA sequences. For identification of Cryptosporidium, most of FISH assays are employed on RNA hybridization rather DNA by making target on a small subunit of nuclear rRNA of variable region [50,54]. Because within cell, small subunit (SSU) has a high copy number considered as rich target. Some limitations with the use of FISH probes depend upon the viability of oocysts as SSU-rRNA decay during cell death making useless the FISH technique. Degradation varies with environmental conditions including RNase contamination, pH, salinity and temperature etc.
- Advanced molecular assays
Different methods are available regarding PCR and many of them have been applied to Cryptosporidium in the past. A variety of techniques have been evaluated, but currently, only those methods are needed which are easy to perform, universally applicable and effective in approach.
- Fingerprinting
Different techniques are available which permit genetic fingerprint to be developed for parasite sample. Such techniques based on genome(s) screening for distinction in organization and sequence. Positive approach of some methods is that no earlier genomic information is needed for a sample which is to be characterized. However, the disadvantage is that pooled organism’s genetic fingerprinting show organism’s population rather than individual, therefore, individual markers may not signify all population. However, these techniques have valuable applications. RAPD (Random Amplification of Polymorphic DNA) or AP-PCR (Arbitrarily Primed-Polymerase Chain Reaction) are developed, based on DNA fragments amplification usually using single primers (10-mer) following separation of amplicons by polyacrylamide or agarose gel electrophoresis. This technique has the benefit of being easy, fast and efficient. Band profiles reproducibility can be raised by using dissimilar thermal cyclers, primer sensitivity, DNA quality and template concentration.
- AFLP (Amplified fragment length polymorphism)
AFLP has been used in parentage analysis, forensic science, for genetic investigation and disease diagnosis in humans and animals. This technique is based on (i) DNA digestion with two restriction enzymes (ii) ligation to 5′ and 3′- end with specific adapters of restriction fragments (iii) use of primers for amplifying restriction fragments (iv) analysis of subsequent restriction fragments through electrophoresis. AFLP has the advantage of being used to composite DNA of every origin. PCR can be applied with high stringency that can attained high discrimination level. Disadvantage of this technique is that it takes more time than other fingerprinting methods. Occasionally, in band profiles a high variability can limit the AFLP performance, but this technique has been utilized to find out the genotype of different organisms, including protozoa but has not been utilized to Cryptosporidium [55]. Some researchers have focused on satellite DNA utilization for genetic make-up analysis of parasite populations. Microsatellites and Minisatellites have been used as ubiquitous and profuse in all eukaryotic genomes. They are usually non-transcribed and sustain polymorphism because of alteration in repeat number. On gel electrophoresis, this change let the alleles to be scored by size and characterization is done by variability in alleles. Thus, it is utilized to investigate genetic mapping and genetic structure of organism’s populations. PCR is an excellent technique for the amplification of repeat region that can be examined by gel electrophoresis and visualized by auto-radiography or staining. Analysis with multi-locus satellite has been used to explore population structures and to identify genetic exchange role in Cryptosporidium [56]. Multi-locus satellite technique has also been utilized to investigate zoonotic threat from several protozoa including Cryptosporidium [57]. Fingerprinting method has also been utilized to explore differences between C. parvum and C. hominis. In electrophoretic band profiles, a similar method revealed the substantial heterogeneity in clinical isolates taken from sporadic diseases and homogeneity among clinical isolates from an outbreak. Based on these studies, it is now clear that these diagnostic tools have given useful information on population structure and diversity in C. hominis and C. parvum and similar methods are required for other Cryptosporidium species.
- PCR-based sequencing and restriction fragment length
Different PCR-based methods, utilizing selective pairs of primers for the amplification of genetic loci followed by sequencing or enzymatic cleavage, have been employed to classify and characterize the Cryptosporidium species genotypes [4]. Several loci (key markers) comprise rRNA genes and spacers, COWP (Cryptosporidium oocyst wall protein), HSP70 (70 kDa heat shock protein), TRAP (Thrombospondin-related adhesive protein) genes and GP60 (genes and the 60 kDa glycoprotein). The SSU-rRNA gene gave a valuable genetic marker for Cryptosporidium identification having great interspecific and low intraspecific sequence differences [7,58]. Markers allow identification at species level include the hsp70 and actin gene which have been used together with SSU (small subunit) in phylogenetic (systematic) analysis of Cryptosporidium, giving basic structure for members classification in the genus [59]. The ITS (internal transcribed spacers) of ribosomal DNA are helpful for revealing variability because their sequences possess great intraspecific variation rather than rRNA gene regions. Extremely variable loci may have repetitive microsatellite regions including gp60, ML1 (microsatellite locus 1) and ML2 (microsatellite locus 2) have been used to explore population genetics of Cryptosporidium especially for C. parvum and C. hominis [60].
- PCR-RFLP (PCR-based restriction fragment length polymorphism)
It has been used to classify Cryptosporidium spp. genetics in many researches. PCR-RFLP based genotypic analysis of gene fragment 18s rRNA has shown 82% isolates from C. hominis, while 18% from C. parvum in children [53]. Moreover, PCR-RFLP analysis for the gene (SSU rRNA) explains that native breeds of cattle do not transmit human Cryptosporidium in Nigeria (Kaduna State). In an advance study, four species of Cryptosporidium, C. bovis, C. andersoni, C. parvum and C. ryanae were explored by analyzing the SSU rRNA (18S) and COWP genes in polish breed cattle. This method does not identify all the sequence length variations in amplicons because endonuclease/s, which is utilized, identifies a limited number of variable sites. One of the gold standard methods remains the direct DNA sequencing to identify polymorphism or genetic variation and can be used for multi-copy (provided there is no sequence heterogeneity in copies) as well as single-copy genes. DNA sequence data can be employed for comparative genetic analysis and suitable for phylogenetic studies. But direct DNA sequencing have some restrictions, as small quantity of DNA from single oocyst of Cryptosporidium is not of practical approach for PCR amplification; amplicons are produced always from that isolate which represents oocysts population. If considerable polymorphism or heterogeneity present in that isolate as in first (ITS-1) and second (ITS-2), ITS regions may not be able to develop a precise sequence from an amplicon [61]. However, analysis of polymorphic amplicons can be done by mutation scanning methods.
- Electrophoretic scanning
Some restrictions in the investigation of sequence variation can be minimized by high resolution electrophoretic methods. These methods are comprised of mutation scanning techniques, for instance, DGGE (denaturing gradient gel electrophoresis), TGGE (temperature gradient gel electrophoresis), SSCP (single strand conformation polymorphism) and hetero duplex analysis are being used. Particularly, SSCP is a valuable method that is based on the principle which states that electrophoretic movement of ssDNA molecule in non-denaturing gel is dependent mainly on two things, its structure and size, enabling the method to verify mutation at a single point in 500bp amplicons [18]. Thus, PCR-based SSCP has exhibited a useful diagnostic tool for identification and genotypes of Cryptosporidium species to verify genetic variation among and within large quantity of samples. In Cryptosporidium, this has also been utilized for exhibiting sequence variation in the regions of nuclear gene (SSU and hsp70) with lesser number of samples from proper oocyst DNA.
New drug discoveries
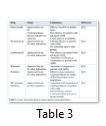
Several therapeutics have been tested against cryptosporidiosis such as macrolide Paromomycin aminoglycoside, ionophores and immunotherapy. However, Nitazoxanide was the only anti-protozoal drug against cryptosporidiosis, which was approved in 2006 by the U.S. food and drug administration. But this drug is only effective in immunocompetent patients. In immunocompromised patients, combination of medicine; paromomycin + protease inhibitors along with antiretroviral therapy is recommended [62-65]. The new drugs recently approved for the treatment of cryptosporidiosis along with their limitations have been mentioned in table 3.
Cryptosporidium spp. is an emerging zoonotic protozoan parasite which is being transmitted through contaminated food, raw vegetables and drinking water which may be a leading cause of gastrointestinal illnesses in humans. Cryptosporidium parvum and Cryptosporidium hominis are most prevalent parasites among all kind of hosts. These species cause chronic diarrhea and dysentery in young children and particularly, in immunocompromised patients or HIV/AIDS patients. Recovery chances from this parasitic infection are very rare in HIV patients. Even, no antiparasitic drug against these virulent protozoa is efficacious in HIV infected/ AIDS patients until a suitable anti-retroviral medicine is given in combination. Nitazoxanide is a drug of choice against Cryptosporidium spp. and is recently approved by FDA. Moreover, the most appropriate option to avoid cryptosporidiosis is to adopt better hygienic and sanitation conditions.
Conflict of Interest Statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
References![]()
- Vanathy K, Parija SC, Mandal J, Hamide A, Krishnamurthy S. Cryptosporidiosis: A mini review. Tropical parasitology, (2017); 7(2): 72.
- Jirku M, Valigurová A, Koudela B, Krízek J, Modrý D, et al. New species of Cryptosporidium Tyzzer, 1907 (Apicomplexa) from amphibian host: morphology, biology and phylogeny. Folia parasitologica, (2008); 55(2): 81.
- Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends in parasitology, (2018).
- Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clinical microbiology reviews, (2004); 17(1): 72-97.
- Kabir M, Ahmed E, Hossain B, Alam M, Ahmed S, et al. Giardia/Cryptosporidium QUIK CHEK® Assay is more Specific than qPCR for Rapid Point-of-care Diagnosis of Cryptosporidiosis in Infants in Bangladesh. Clinical Infectious Diseases, (2018); 67(12): 1897-1903.
- Checkley W, White Jr AC, Jaganath D, Arrowood MJ, Chalmers RM, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. The Lancet Infectious Diseases, (2015); 15(1): 85-94.
- Fayer R. Taxonomy and species delimitation in Cryptosporidium. Experimental parasitology, (2010); 124(1): 90-97.
- Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, et al. Molecular characterization of Cryptosporidium spp. in HIV-infected persons in Benin City, Edo State, Nigeria. Fooyin Journal of Health Sciences, (2010); 2(3-4): 85-89.
- Abdel-Messih IA, Wierzba TF, Abu-Elyazeed R, Ibrahim AF, Ahmed SF, et al. Diarrhea associated with Cryptosporidium parvum among young children of the Nile River Delta in Egypt. Journal of Tropical Pediatrics, (2005); 51(3): 154-159.
- Mbae CK, Nokes DJ, Mulinge E, Nyambura J, Waruru A, et al. Intestinal parasitic infections in children presenting with diarrhoea in outpatient and inpatient settings in an informal settlement of Nairobi, Kenya. BMC Infectious Diseases, (2013); 13(1): 243.
- Delahoy MJ, Omore R, Ayers TL, Schilling KA, Blackstock AJ, et al. Clinical, environmental, and behavioral characteristics associated with Cryptosporidium infection among children with moderate-to-severe diarrhea in rural western Kenya, 2008–2012: The Global Enteric Multicenter Study (GEMS). PLoS neglected tropical diseases, (2018); 12(7): e0006640.
- Goma F, Geurden T, Siwila J, Phiri I, Gabriël S, et al. The prevalence and molecular characterisation of Cryptosporidium spp. in small ruminants in Zambia. Small Ruminant Research, (2007); 72(1): 77-80.
- Mensah G, Bosompem K, Ayeh-Kumi P, Oppong J, Niampoma S. Epidemiology of Cryptosporidium Sp Upstream the Water Treatment Plants in Kpong and Weija, Ghana. Ghana Journal of Science, (2018); 585-11.
- Marques SMT. Cryptosporidiosis in Horses of Urban Areas of Porto Alegre, Rio Grande do Sul, Southern Brazil. Journal of equine veterinary science, (2010); 30(7): 356-358.
- Iqbal A, Goldfarb DM, Slinger R, Dixon BR. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in diarrhoeic patients in the Qikiqtani Region, Nunavut, Canada. International journal of circumpolar health, (2015); 74(1): 27713.
- Gatei W, Barrett D, Lindo JF, Eldemire-Shearer D, Cama V, et al. Unique Cryptosporidium population in HIV-infected persons, Jamaica. Emerging Infectious Diseases, (2008); 14(5): 841.
- Alonso‐Fresan M, Garcia‐Alvarez A, Salazar‐Garcia F, Vázquez‐Chagoyán J, Pescador‐Salas N, et al. Prevalence of Cryptosporidium spp. in asymptomatic sheep in family flocks from Mexico State. Journal of Veterinary Medicine, Series B, (2005); 52(10): 482-483.
- Giangaspero A, Iorio R, Paoletti B, Traversa D, Capelli G. Molecular evidence for Cryptosporidium infection in dogs in Central Italy. Parasitology Research, (2006); 99(3): 297-299.
- Causape A, Quilez J, Sanchez-Acedo C, Del Cacho E. Prevalence of intestinal parasites, including Cryptosporidium parvum, in dogs in Zaragoza city, Spain. Veterinary parasitology, (1996); 67(3-4): 161-167.
- Quilez J, Sanchez-Acedo C, Del Cacho E, Clavel A, Causape A. Prevalence of Cryptosporidium and Giardia infections in cattle in Aragon (northeastern Spain). Veterinary Parasitology, (1996); 66(3-4): 139-146.
- Chilvers B, Cowan P, Waddington D, Kelly P, Brown T. The prevalence of infection of Giardia spp. and Cryptosporidium spp. in wild animals on farmland, southeastern North Island, New Zealand. International Journal of Environmental Health Research, (1998); 8(1): 59-64.
- Winkworth C, Matthaei C, Townsend C. Prevalence of Giardia and Cryptosporidium spp in calves from a region in New Zealand experiencing intensifi cation of dairying. New Zealand veterinary journal, (2008); 56(1): 15-20.
- Atwill ER, Sweitzer RA, Pereira M, Gardner IA, Van Vuren D, et al. Prevalence of and associated risk factors for shedding Cryptosporidium parvum oocysts and Giardia cysts within feral pig populations in California. Applied and Environmental Microbiology, (1997); 63(10): 3946-3949.
- Wang A, Ruch-Gallie R, Scorza V, Lin P, Lappin MR. Prevalence of Giardia and Cryptosporidium species in dog park attending dogs compared to non-dog park attending dogs in one region of Colorado. Veterinary Parasitology, (2012); 184(2-4): 335-340.
- Leach CT, Koo FC, Kuhls TL, Hilsenbeck SG, Jenson HB. Prevalence of Cryptosporidium parvum infection in children along the Texas-Mexico border and associated risk factors. The American journal of tropical medicine and hygiene, (2000); 62(5): 656-661.
- Rickard LG, Siefker C, Boyle CR, Gentz EJ. The prevalence of Cryptosporidium and Giardia spp. in fecal samples from free-ranging white-tailed deer (Odocoileus virginianus) in the southeastern United States. Journal of Veterinary Diagnostic Investigation, (1999); 11(1): 65-72.
- Petri Jr WA, Haque R, Mondal D, Karim A, Molla IH, et al. Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clinical Infectious Diseases, (2009); 48(9): 1191-1197.
- Mallinath HK, G Chikkachowdappa P, K Javare Gowda A, E D’Souza P. Studies on the prevalence of cryptosporidiosis in bovines in organized dairy farms in and around Bangalore, South India. Veterinarski arhiv, (2009); 79(5): 461-470.
- Bhat S, Juyal P, Singla L. Prevalence of cryptosporidiosis in neonatal buffalo calves in Ludhiana district of Punjab, India. Asian J Anim Vet Adv, (2012); 7(6): 512-520.
- Ghoshal U, Jain V, Dey A, Ranjan P. Evaluation of enzyme linked immunosorbent assay for stool antigen detection for the diagnosis of cryptosporidiosis among HIV negative immunocompromised patients in a tertiary care hospital of northern India. Journal of infection and public health, (2018); 11(1): 115-119.
- Rahi AA, Magda A, Al-Charrakh AH. Prevalence of Cryptosporidium parvum among children in Iraq. American Journal of Life Sciences, (2013); 1(6): 256-260.
- Butty E. Detection of Cryptosporidium and Giardia doudenalis in equines in Nineveh, Iraq. Iraqi Journal of Veterinary Sciences, (2011); 25(2): 43-46.
- Ahamed I, Yadav A, Katoch R, Godara R, Saleem T, et al. Prevalence and analysis of associated risk factors for Cryptosporidium infection in lambs in Jammu district. Journal of parasitic diseases, (2015); 39(3): 414-417.
- Ghimire P, Sapkota D, Manandhar SP. Cryptosporidiosis: opportunistic infection in HIV/AIDS patients in Nepal. J Trop Med Parasitol, (2004); 277-10.
- Nasir A, Avais M, Khan M, Ahmad N. Prevalence of Cryptosporidium parvum infection in Lahore (Pakistan) and its association with diarrhea in dairy calves. Int J Agric Biol, (2009); 11(2): 221-224.
- Mumtaz S, Ahmed J, Ali L. Frequency of cryptosporidium infection in children under five years of age having diarrhea in the North West of Pakistan. African Journal of Biotechnology, (2010); 9(8); 1230-1235.
- Srisuphanunt M, Karanis P, Charoenca N, Boonkhao N, Ongerth JE. Cryptosporidium and Giardia detection in environmental waters of southwest coastal areas of Thailand. Parasitology research, (2010); 106(6): 1299-1306.
- Waldron LS, Dimeski B, Beggs PJ, Ferrari BC, Power ML. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Applied and environmental microbiology, (2011); 77(21): 7757-7765.
- Power M, Shanker S, Sangster N, Veal D. Evaluation of a combined immunomagnetic separation/flow cytometry technique for epidemiological investigations of Cryptosporidium in domestic and Australian native animals. Veterinary parasitology, (2003); 112(1-2): 21-31.
- Ryan U, Samarasinghe B, Read C, Buddle J, Robertson I, et al. Identification of a novel Cryptosporidium genotype in pigs. Applied and Environmental Microbiology, (2003); 69(7): 3970-3974.
- Karanis P, Kourenti C, Smith H. Waterborne transmission of protozoan parasites: a worldwide review of outbreaks and lessons learnt. Journal of water and health, (2007); 5(1): 1-38.
- Thompson RA, Colwell DD, Shury T, Appelbee AJ, Read C, et al. The molecular epidemiology of Cryptosporidium and Giardia infections in coyotes from Alberta, Canada, and observations on some cohabiting parasites. Veterinary parasitology, (2009); 159(2): 167-170.
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science, (2004); 304(5669): 441-445.
- Muchiri JM, Ascolillo L, Mugambi M, Mutwiri T, Ward HD, et al. Seasonality of Cryptosporidium oocyst detection in surface waters of Meru, Kenya as determined by two isolation methods followed by PCR. Journal of water and health, (2009); 7(1): 67-75.
- Bomfim T, Huber F, Gomes R, Alves L. Natural infection by Giardia sp. and Cryptosporidium sp. in dairy goats, associated with possible risk factors of the studied properties. Veterinary parasitology, (2005); 134(1-2): 9-13.
- El-Khodery SA, Osman SA. Cryptosporidiosis in buffalo calves (Bubalus bubalis): prevalence and potential risk factors. Tropical animal health and production, (2008); 40(6): 419-426.
- Tian L-G, Chen J-X, Wang T-P, Cheng G-J, Steinmann P, et al. Co-infection of HIV and intestinal parasites in rural area of China. Parasites & vectors, (2012); 5(1): 36.
- Adjei AA, Armah H, Rodrigues O, Renner L, Borketey P, et al. Cryptosporidium spp., a frequent cause of diarrhea among children at the Korle-Bu Teaching Hospital, Accra, Ghana. Japanese journal of infectious diseases, (2004); 57(5): 216-219.
- Wumba R, Longo-Mbenza B, Menotti J, Mandina M, Kintoki F, et al. Epidemiology, clinical, immune, and molecular profiles of microsporidiosis and cryptosporidiosis among HIV/AIDS patients. International journal of general medicine, (2012); 5603.
- Smith H (2007) Diagnostics. Cryptosporidium and Cryptosporidiosis, Second Edition: CRC Press. pp. 181-215.
- Smith H, Caccio S, Cook N, Nichols R, Tait A. Cryptosporidium and Giardia as foodborne zoonoses. Veterinary parasitology, (2007); 149(1-2): 29-40.
- Uppal B, Singh O, Chadha S, Jha AK. A comparison of nested PCR assay with conventional techniques for diagnosis of intestinal cryptosporidiosis in AIDS cases from northern India. Journal of parasitology research, (2014); 2014.
- Nyamwange CI (2013) Epidemiology and Molecular Characterization of Cryptosporidium Species among Children and HIV Infected Individuals in the North Rift Region of Kenya. PhD Thesis. Jomo Kenyatta University of Agriculture and Technology
- Bednarska M, Bajer A, Sinski E, Girouard AS, Tamang L, et al. Fluorescent in situ hybridization as a tool to retrospectively identify Cryptosporidium parvum and Giardia lamblia in samples from terrestrial mammalian wildlife. Parasitology research, (2007); 100(3): 455-460.
- Blake D, Smith A, Shirley M. Amplified fragment length polymorphism analyses of Eimeria spp.: an improved process for genetic studies of recombinant parasites. Parasitology research, (2003); 90(6): 473-475.
- Mallon M, MacLeod A, Wastling J, Smith H, Reilly B, et al. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. Journal of Molecular Evolution, (2003); 56(4): 407-417.
- Grinberg A, Lopez-Villalobos N, Pomroy W, Widmer G, Smith H, et al. Host-shaped segregation of the Cryptosporidium parvum multilocus genotype repertoire. Epidemiology & Infection, (2008); 136(2): 273-278.
- Jex AR, Whipp M, Campbell BE, Cacciò SM, Stevens M, et al. A practical and cost‐effective mutation scanning‐based approach for investigating genetic variation in Cryptosporidium. Electrophoresis, (2007); 28(21): 3875-3883.
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Experimental parasitology, (2010); 124(1): 80-89.
- Chalmers RM, Katzer F. Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends in parasitology, (2013); 29(5): 237-251.
- Schindler AR, EL-Osta YGA, Stevens M, Sinclair MI, Gasser RB. Capillary electrophoretic analysis of fragment length polymorphism in ribosomal markers of Cryptosporidium from humans. Molecular and cellular probes, (2005); 19(6): 394-399.
- Cabada MM, White Jr AC. Treatment of cryptosporidiosis: do we know what we think we know? Current opinion in infectious diseases, (2010); 23(5): 494-499.
- Rossignol JF, Kabil SM, El–Gohary Y, Younis AM. Effect of nitazoxanide in diarrhea and enteritis caused by Cryptosporidium species. Clinical Gastroenterology and Hepatology, (2006); 4(3): 320-324.
- Wang RJ, Li JQ, Chen YC, Zhang LX, Xiao LH. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: Epidemiology, clinical feature, diagnosis, and therapy. Acta tropica, (2018); 187:257-263.
- Waywa D, Silpasakorn S, Phungthaisong A, Suputtamongkol Y. Intestinal Parasitic Infections in Thai Patients: Five-Year Experiences at Siriraj Hospital. Siriraj Medical Journal, (2018); 58(11): 1107-1109.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0