Full Length Research Article
Nitrogen removal efficiency of some bacterial strains isolated from seawater in Thua Thien Hue, Vietnam
Le Cong Tuan1, Nguyen Duc Huy2, Le My Tieu Ngoc2, Doan Thi My Lanh1, Te Minh Son1, Nguyen Hoang Loc3*
Adv. life sci., vol. 8, no. 2, pp. 184-189, February 2021
*- Corresponding Author: Nguyen Hoang Loc (Email: nhloc@hueuni.edu.vn)
Authors' Affiliations
2. Institute of Biotechnology, Hue University, Thua Thien Hue – Vietnam
3. Institute of Bioactive Compounds, University of Sciences, Hue University, Hue 530000 – Vietnam
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Nitrifying bacteria in aquaculture environments are capable of removing toxic nitrogen compounds such as ammonium and nitrite. Using these indigenous microbial resources can improve shrimp production.
Methods: Screening method was used to isolate aerobic strains of nitrifying bacteria. Species identification for these isolates was done by biomolecular method based on 16S rDNA gene sequence. Ammonium, nitrite and nitrate concentrations from the culture were determined by spectrophotometry at the appropriate wavelength. Temperature, pH, dissolved oxygen and salinity were measured by specialized equipment. Formation and development of flocs during shrimp culture were determined based on their volume and weight. A trial of shrimp nursery was carried out on a small scale with 0.5 m3 tanks containing diluted seawater to 16-18‰ salinity at a density of 400 individual/m3 for 24 days on April 2019.
Results: This study isolated two strains of Pseudomonas (BF01 and BF03) and one strain of Cupriavidus oxalaticus BF02 from seawater in Thua Thien Hue province, Vietnam. These bacterial isolates have shown ability to remove nitrogen compounds such as ammonium, nitrite and nitrate in culture medium. Formation and development of flocs were found in trials of shrimp nursery with diluted seawater containing the isolates. Some water quality parameters (temperature, pH, dissolved oxygen, salinity, ammonium and nitrite) were kept at a safe level and juvenile shrimp grown normally during culture.
Conclusion: The observations on the water quality and basic growth parameters of juvenile shrimp in the two treatments, diluted seawater and diluted seawater with commercial microbial products, showed that there were no significant differences between them with p = 0.05. This proves that three isolates have played an important role in shrimp nursery.
Keywords: Cupriavidus oxalaticus; Floc; Litopenaeus vanamei; Nitrifying-denitrifying bacteria; Pseudomonas sp.
In aquaculture, shrimp farming is considered to be one of the important industries accounting for about four million tons of output worldwide each year. However, the productivity of shrimp depends on many factors such as microbial community, oxygen and nitrogen concentration, temperature and waste composition in the aquatic environment [1]. Among them, nitrogen concentration plays a key role and in fact, nitrogen sources including nitrite and ammonia are two important indicators of water quality not only for shrimp but also for other aquatic animals [2].
Ammonium appears as a product of the conversion of organic waste in aquatic animals and the breakdown of food by microorganisms [3]. Whereas, nitrite is accumulated from the action of nitrifying bacteria or the ammonia oxidation [4]. These two substance groups directly impact on the productivity of aquaculture [2]. Therefore, low concentration of ammonium and nitrite should be maintained during farming. The management of ammonium and nitrite must be carried out regularly by water exchange. However, recent reports have shown that water exchange without treatment can pollute the environment, increasing pathogenic activity of microorganisms [2, 5, 6]. As a result, new technologies have been developed to treat water before being released into the environment, such as recirculating aquaculture system and biological flocs co-culture or biofloc technology (BFT) [7-10]. The BFT was considered the new “blue revolution” in aquaculture, this technology is an environmentally friendly aquaculture procedure based on in situ microbial production. Fish and shrimp are usually grown in an intensive way with zero or minimum water exchange. In addition, continuously water movement in the entirely water column is also required to induce the macroaggregate (biofloc) formation [11]. BFT enhances water quality by converting organic matter into microbial biomass, reducing feed intake by using natural feed in the culture system, improving the health and productivity of farmed shrimp, and reducing negative impacts of aquaculture on the environment [8, 9]. In the biofloc-based system, ammonium and nitrite are removed through nitrification by ammonia-oxidizing bacteria and subsequently nitrite-oxidizing bacteria. Thus, heterotrophic nitrifying bacteria and ammonium assimilation bacteria have attracted great interest in the field of aquaculture science [9, 12-14].
In the present work, we report some results of nitrogen removal efficiency of three bacterial strains isolated from seawater in Thua Thien Hue province, Vietnam. The ability to remove ammonium and nitrite in a small-scale juvenile shrimp culture environment of these native isolates opens up their potential applications in large-scale shrimp production in the future.
Screening of aerobic nitrifying-denitrifying bacteria
Enrichment and screening of nitrifying-denitrifying bacteria were conducted as described by Qiu et al [15] with slight modifications. 100 µL of seawater samples in Thua Thien Hue, Vietnam were added to 50 mL of sterilized enrichment medium containing 0.5 g/L ammonium sulfate, 2.17 g/L sodium succinate and 50 mL/L mineral solution (per liter: 5 g dipotassium phosphate, 2.5 g magnesium sulfate heptahydrate, 2.5 g sodium chloride, 0.05 g ferrous sulfate heptahydrate and 0.05 g manganese (II) sulfate tetrahydrate, pH 7.2) and incubated at 30°C on shaker with a speed of 180 rpm for 7 days. Then, 100 µL of enrichment culture were plated on selection medium (per liter: 1 g potassium nitrate, 1 g monopotassium phosphate, 0.5 g ferrous sulfate heptahydrate, 0.2 g calcium chloride, 1 g magnesium sulfate heptahydrate, 8.5 g sodium succinate, 20 g agar and 1 mL bromothymol blue (1% in alcohol, w/v), pH 7-7.3) and incubated at 30°C for 7 days. Blue colonies appeared on agar surface were selected for investigation of their nitrogen conversion.
Nitrogen conversion of isolated bacteria
Blue colonies were subcultured in 5 mL of medium containing (per liter) 0.84 g sodium nitrate, 1 g monopotassium phosphate, 1 g magnesium sulfate heptahydrate, 4.16 g glucose, 0.05 g ferrous sulfate heptahydrate and 0.02 g calcium chloride, pH 7 and incubated at 30oC with a shaking speed of 180 rpm for 24 h. Culture was then transferred into new media consists of (per liter): 1) 3.38 g sodium succinate, 0.33 g ammonium sulfate, 0.1 g peptone, 0.08 g sodium chloride, 0.023 g calcium carbonate, 0.023 g mono potassium phosphate, 0.094 g magnesium sulfate , 0.065 g sodium bicarbonate, 0.2 mg ferrous sulfate heptahydrate, 0.2 mg manganese sulfate monohydrate, 0.2 mg copper sulfate pentahydrate and 0.2 g cobalt (II) chloride hexahydrate (for nitrification); and 2) 1 g mono potassium phosphate, 1 g magnesium sulfate heptahydrate, 2.8 g sodium succinate and 0.6 g sodium nitrate (for denitrification) or 0.5 g sodium nitrite (for nitrification). Nitrogen conversion of isolates was determined based on the changes of nitrogen concentration in the culture medium that measured by the standard methods [16].
Molecular identification
Bacterial biomass was harvested from overnight culture in 5 mL of Luria-Bertani (LB) medium at 30°C and 180 rpm. Total DNA from bacterial biomass was isolated by AquaPure Genomic Isolation Kit (732-6340, Bio-Rad) following the manufacturer’s instructions. Identification of the bacteria was performed by sequencing 16S rDNA gene. Universal primers were used for amplification of 16S region (27F: 5’-AGAGTTTGATCCTGGCTCAG-3’ and 1492R: 5’-GGTTACCTTGTTACGACTT-3’). Polymerase chain reaction (PCR) component consists of 6 µL of Master Mix (Thermo Fisher Scientific), 10 pmol each primer, 50 ng of total DNA and double distilled water to a final volume of 12 µL. PCR amplification was done as follows: a genomic denaturation at 95ºC for 5 min; 30 cycles of 95ºC for 1 min, 55ºC for 1 min, and 72ºC for 1.5 min; and a final extension at 72ºC for 10 min. The amplicon was sequenced by the dideoxy chain termination method on the Applied Biosystem 3130 (ABI). Phylogenetic tree of 16S rDNA gene was constructed by MEGA X software with the Maximum Likelihood algorithm, bootstrap of 500 [17].
Shrimp farming trial
Experiments were distributed on a completely randomized design, with two treatments (DS: diluted seawater and DSM: diluted seawater with microbial product) and three replicates per treatment. Disease-free post larvae 12 (10 mg/individual) of Pacific white shrimp (Litopenaeus vannamei) were cultured in six composite tanks (volume of 0.5 m3 each tank) containing seawater diluted to 16-18‰ salinity from natural seawater (30‰ salinity) and sterilized distilled water [18] at a density of 400 individual/m3 for 24 days (April, 2019) .
Molasses sugar was added to culture tank in a carbon/nitrogen ratio of 15/1 to allow microorganisms to form flocs [19]. The aeration was done with a speed of 3.7 m3/min (Dargang 2HP DG-400-31) and the bottom mud was stirred at water flow of 3.9 m3/h by Lifetech AP 5400 pump. For DSM treatment, weekly 1 g of commercial microbial product containing Nitrobacter spp. (6.85×105 CFU/g), Nitrosomonas spp. (3.95×105 CFU/g) and Bacillus subtilis (3.37×107 CFU/g) from ALT Co., Ltd. (Vietnam) were added to culture tanks as the control.
Shrimp food was purchased from Grobest Industrial Co., Ltd. (Vietnam) containing 42% protein. Shrimps were fed 3 times a day at 7:00, 12:00 and 17:00. Feed amount was calculated following formula:
y = 13.391×W-0.5558, where y is the amount of feed provided and W is the weight of shrimp [20]. During shrimp culture, no antibiotics or any other substances were used; water quality parameters such as temperature, pH, dissolved oxygen (DO), salinity, NH4-N, and NO2-N, floc dry weight and volume were determined every 4-5 days at 10:00. Shrimp performance was assessed at the end of the experiment based on the following parameters: growth rate (GR) by weight, growth rate by size, survival rate and feed conversion ratio (FCR is the total weight of harvested shrimp divided by the total amount of feed used).
Determination of water quality parameters
Water quality parameters during shrimp culture were measured by HI 9142 portable waterproof (HANNA) for temperature and DO, pH100A (EcoSense) for pH, and Sension 156 portable multiparameter meter (HACH) for salinity. 100 mL water from the culture after filtration with Whatman GF/F filter paper to be used to determine concentration of ammonium and nitrite by the optical measurement method on Shimadzu UV1800 spectrophotometer. Determination of ammonium based on the indophenol reaction with o-phenylphenol (OPP) [21]. Nitrate is reduced almost quantitatively to nitrite in the presence of Cd. The nitrite thus produced is determined by diazotizing with sulfanilamide and coupling with N-(1-naphthy)-ethylenediamine dihydrochloride to form a highly colored azo dye that is measured colorimetrically [16].
Determination of floc growth
Floc volume (mL/L) in the culture tanks was measured by Imhoff cones.
Fresh weight (FW, g/L) of floc (mg/L) was harvested at 7:00 and dry to a constant weight at 105°C. Fresh and dry weight (DW, g/L) were determined by the weighting method following formulas:
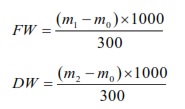
Statistical analysis
All experiments were preformed with three replications per treatment. The data are represented as the mean of the repeats; the means were compared using one-way ANOVA (Duncan’s test at significance level of 0.05).
Screening and identification of bacteria
Three isolates of aerobic nitrifying-denitrifying bacteria obtained from seawater samples in Thua Thien Hue, Vietnam were named BF01, BF02 and BF03 (Figure 1). Molecular identifications for these isolates were done and their 16S gene sequences were deposited in the Genbank (NCBI) with accession numbers: MN559954 for Pseudomonas sp. BF01, MN559955 for Cupriavidus oxalaticus BF02, and MN559956 for Pseudomonas sp. BF03. Phylogenetic tree of C. oxalaticus BF02 and other Cupriavidus species was constructed as showed in Figure 2, of which 16S gene sequences of C. oxalaticus BF02 strain is 99.93% homologous with that of C. oxalaticus X32 (accession number in NCBI: MG754065.2).
Nitrogen conversion of bacteria
Nitrogen conversion of isolated bacteria was investigated and data were showed in Tables 1-3. Table 1 shows that nitrification has occurred relatively strongly, 28 to 46.7% of the ammonium in the medium was removed by isolates, of which C. oxalaticus BF02 displayed the highest ability. Although nitrite could not be detected in the medium, nitrate formation was found in the range of 2.3 to 5.1 mg/L. In second step of nitrification, 2.2 to 15% of the nitrite was removed and the nitrate was found from 0.3 to 11.47 mg/L (Table 2). The results in Table 3 indicate that denitrification was strongly conducted by two isolates of Pseudomonas sp. BF01 and C. oxalaticus BF02 with 55.6 to 75.8% of nitrate in the medium was removed. However, this ability of Pseudomonas sp. BF03 seems insignificant, only about 6%. Nitrite formation was not found in this process.
Overall, C. oxalaticus BF02 isolate exhibited the stronger nitrogen conversion than the other two isolates, Pseudomonas BF01 and BF03.
Shrimp farming trial in small-scale
Investigation of physical-chemical parameters such as temperature, pH, DO and salinity in both treatments, DS and DSM, during shrimp culture showed they were relatively stable and ranged around 30°C, 7.5, 7.4 and 16.9, respectively. The differences between corresponding parameters were statistically insignificant with p>0.05 (Table 4). In general, the values of these parameters are suitable for shrimp farming [22].
Ammonium concentration in both treatments increased from the beginning to 15th day of culture with maximum values of 1.19 and 1.27 mg/L, but nitrite alone increased continuously until the 24th day to 2.31 and 2.55 mg/L. However, ammonium then decreased significantly from day 15, to only about 0.52-0.69 mg/L (Figure 3 and 4). Low concentrations of ammonium and nitrite in this investigation, peaked respectively only 1.27 mg/L (equivalent to 2.34 mg/L of ammonia-N or 0.11 mg/L of NH3-N) and 2.55 mg/L at about 16.9‰ salinity (Figures 3 and 4, and Table 4) during culture, did not adversely affect shrimp growth.
Figures 5 and 6 show floc dry weight and volume increased continuously with time during shrimp culture with values of approximately 46 mg/L (control was 43 mg/L) and 10 mL/L (control was 8 mL/L), respectively. Differences of floc dry weight and volume in both treatments, DS and DSM, were statistically significant with p<0.05.
Some growth parameters of juvenile shrimp are shown in Table 5. Growth rate (GR) of shrimp by weight and size averaged 25.5 mg/day (control was 25.02 mg/day) and 0.17 cm/day (control was 0.21 cm/day), respectively. Survival rates and FCRs of shrimp in both treatments were similar, approximately 82.5% and 0.8, respectively. In general, differences of growth parameters of juvenile shrimp between DS and DSM treatments were statistically insignificant with p>0.05.
Figures & Tables
Many previous reports indicated that Pseudomonas bacteria has a strong ability to convert nitrogen [22-26]. However, most studies showed that C. oxalaticus bacteria were only able to metabolize hazardous organic compounds such as acrylamide [27] or metal ions as cadmium [28] or others as 2, 6-dibromo-4-nitrophenol [29]. Except for a recent report of nitrifying and denitrifying ability of this bacterium [30] and our findings thus reinforced that study.
In general, three isolated bacterial strains can biologically oxidize ammonium to nitrite, the reaction usually was represented by the Nitrosomonas bacteria; or convert nitrite to nitrate, the reaction usually was represented by the Nitrobacter bacteria [31]. The results in Table 1 suggested that most of the nitrite formed from ammonium may have been converted into nitrate [32], and bacteria can use nitrate compounds as a nutrient for growth [33]. Data from Table 2 show that the isolates converted nitrite to nitrate, a nitrogen compound less toxic than nitrite and ammonium [34, 35]. In addition, in our opinion the reason for the absence of nitrite in the medium (Table 3) is because nitrates have been used by bacteria as a source of nutrition. In conclusion, these isolates can be applied to minimize ammonium and nitrite contamination in shrimp culture environment.
In shrimp farming trials, an increase in ammonium concentration during the first 15 days may be due to the nitrogen compounds in the faeces that were excreted by the shrimp as metabolic waste or from shrimp feed [36, 37]. However, microorganisms in flocs, including three isolates and two Bacillus isolates (data not shown), have been involved in ammonium conversion during shrimp culture resulting in a reduced concentration of this substance (Figure 2). The results in Table 2 show that the nitrite removal of three isolates was relatively weak so the amount of nitrite has increased continuously during shrimp culture (Figure 3), but it was still below the allowed safety threshold. Lin and Chen [38] suggested that a safe concentration of nitrite-N for rearing L. vannamei juveniles at 15‰ and 25‰ salinity to be 6.1 mg/L and 15.2 mg/L. Also according to Lin and Chen [39], the “safety level” of ammonia-N or NH3-N for rearing L. vannamei juveniles was estimated to be 2.44 mg/L and 3.55mg/L or 0.12 mg/L and 0.16 mg/L at 15‰ and 25 ‰ salinity, respectively.
In conclusion, the addition of commercial microbial product to the shrimp culture environment did not make a significant difference in water quality parameters and basic growth parameters of juvenile shrimp. This proves that three isolates have played an important role in shrimp nursery.
Author Contributions
Loc NH: Study design, Processed experimental data and analyzed the results, wrote the manuscript.
Tuan LC: Conducted the research work, processed experimental data and analyzed the results.
Ngoc LMT, Lanh DTM, Son TM, Huy ND: Conducted the research work.
This study was supported by the grant of Vietnam Ministry of Education and Training (Code: B2019-DHH-08). The authors would also like to thank Hue University, Vietnam for supporting this study.
- Chen YH, He JG. Effects of environmental stress on shrimp innate immunity and white spot syndrome virus infection. Fish & Shellfish Immunology, (2019); 84: 744-755.
- Yun L, Yu Z, Li Y, Luo P, Jiang X, et al. Ammonia nitrogen and nitrite removal by a heterotrophic Sphingomonas sp. strain LPN080 and its potential application in aquaculture. Aquaculture, (2019); 500: 477-484.
- Gross A, Abutbul S, Zilberg D. Acute and chronic effects of nitrite on white shrimp, Litopenaeus vannamei, cultured in low-salinity brackish water. Journal of the World Aquaculture Society, (2004); 35: 315-321.
- Wang WN, Wang AL, Zhang Y, Li ZH, Wang JX, et al., Effects of nitrite on lethal and immune response of Macrobrachium nipponense. Aquaculture, (2004); 232: 679-686.
- Kautsky N, Rönnbäck P, Tedengren M, Troell M. Ecosystem perspectives on management of disease in shrimp pond farming. Aquaculture, (2000): 191: 145-161.
- Ferreira NC, Bonetti C, Seifferta WQ. Hydrological and water quality indices as management tools in marine shrimp culture. Aquaculture, (2011); 318: 425-433.
- Brown MN, Briones A, Diana J, Raskin L. Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiology Ecology, (2013); 83: 17-25.
- Bossier P, Ekasari J. Biofloc technology application in aquaculture to support sustainable development goals. Microbial Biotechnology, (2017); 10: 1012-1016.
- Santhana Kumar V, Pandey PK, Anand T, Bhuvaneswari GR, Dhinakaran A, et al. Biofloc improves water, effluent quality and growth parameters of Penaeus vannamei in an intensive culture system. Journal of Environmental Management, (2018); 215: 206-215.
- Chen Z, Chang Z, Zhang L, Jiang Y, Ge H, et al. Effects of water recirculation rate on the microbial community and water quality in relation to the growth and survival of white shrimp (Litopenaeus vannamei). BMC Microbiology, (2019); 19: 192.
- Emerenciano MGC, Martínez-Córdova LR, Martínez-Porchas M, Miranda-Baeza A. Biofloc Technology (BFT): Tool for Water Quality Management in Aquaculture. Chapter 5. In: Water Quality (Tutu H, ed.), (2017); IntechOpen Ltd., London.
- Xu WJ, Pan LQ. Enhancement of immune response and antioxidant status of Litopenaeus vannamei juvenile in biofloc-based culture tanks manipulating high C/N ratio of feed input. Aquaculture, (2013); 412-413: 117-124.
- Ruan Y, Taherzadeh MJ, Kong D, Lu H, Zhao H, et al. Nitrogen removal performance and metabolic pathways analysis of a novel aerobic denitrifying halotolerant Pseudomonas balearica strain RAD-17. Microorganisms, (2020); 8: 1.
- Luo L, Zhao Z, Huang X, Du X, Wang C, et al. Isolation, identification, and optimization of culture conditions of a bioflocculant-producing bacterium Bacillus megaterium SP1 and its application in aquaculture wastewater treatment. BioMed Research International, (2016); 2016: 2758168.
- Qiu X, Wang T, Zhong X, Du G, Chen J. Screening and characterization of an aerobic nitrifying-denitrifying bacterium from activated sludge. Biotechnology and Bioprocess Engineering, (2012); 17: 353-360.
- Baird RB, Eaton AD, Rice EW. Standard Methods for the Examination of Water and Wastewater. 23rd ed., (2017); American Public Health Association, American Water Works Association, and Water Environment Federation, USA.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, (2018); 35: 1547-1549.
- Boyd CE. Water Quality Management and Aeration in Shrimp Farming. 2nd ed., (1989); Fisheries and Allied Aquacultures Departmental Series. Alabama Agricultural Experiment Station, USA.
- Avnimelech Y. Carbon/nitrogen ratio as a control element in aquaculture systems. Aquaculture, (1999); 176: 227-235.
- Wyk PV, Samocha TM, David AD, Lawrence AL, Collins CR. Intensive and super-intensive production of the Pacific White leg (Litopenaeus vannamei) in greenhouse-enclose raceway system. In Book of Abstracts, Aquaculture 2001, Lake Buena Visa, L, 573P.
- Kanda J. Determination of ammonium in seawater based on the indophenol reaction with o- phenylphenol (OPP). Water Research, (1995); 29: 2746-2750.
- Xu Y, He T, Li Z, Ye Q, Chen Y. Nitrogen removal characteristics of Pseudomonas putida Y-9 capable of heterotrophic nitrification and aerobic denitrification at low temperature. BioMed Research International, (2017); 1429018.
- Fitriyanto NA, Winarti A, Imara FA, Erwanto Y, Hayakawa T. Identification and growth characters of nitrifying Pseudomonas sp., LS3K isolated from Odorous region of Poultry farm. Journal of Biological Sciences, (2017); 17: 1-10.
- Zhang J, Wu P, Hao B, Yu Z. Heterotrophic nitrification and aerobic denitrification by the bacterium Pseudomonas stutzeri YZN-001. Bioresource Technology, (2011); 102: 9866-9869
- Prasetyo RA, Pertiwiningrum A, Erwanto Y, Yusiati LM, Fitriyanto NA. The potency of Pseudomonas sp. LS3K as nitrifying bacteria on inorganic medium at various c/n ratios. Asian Journal of Microbiology, Biotechnology, Environmental Sciences, (2019); 21: 257-263.
- Zhou M, Ye H, Zhao X. Isolation and characterization of a novel heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas stutzeri KTB for bioremediation of wastewater. Biotechnology Bioprocess Engineering, (2014); 19: 231-238.
- Bedade DK, Singhal RS. Biodegradation of acrylamide by a novel isolate, Cupriavidus oxalaticus ICTDB921: Identification and characterization of the acrylamidase produced. Bioresource Technology, (2018); 261: 122-132.
- Su JF, Xue L, Huang TL, Wang Z, Wang JX. Kinetic analysis of denitrification coupled with Cd(II) removal by Cupriavidus sp. CC1 and its removal mechanism. Research in Microbiology, (2019); 170: 214-221.
- Min J, Chen W, Hu X. Biodegradation of 2,6-dibromo-4-nitrophenol by Cupriavidus sp. strain CNP-8: Kinetics, pathway, genetic and biochemical characterization. Journal of Hazardous Materials, (2019); 361: 10-18.
- Sun Z, Lv Y, Liu Y, Ren R. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a novel metal resistant bacterium Cupriavidus sp. S1. Bioresource Technology, (2016); 220: 142-150.
- Ferguson SJ, Richardson DJ, Van Spanning RJM. Biochemistry and Molecular Biology of Nitrification. In: Biology of the Nitrogen Cycle (Bothe H, Ferguson SJ, Newton WE, eds.), (2007); Elsevier Science, Amsterdam, The Netherlands.
- Tachiki T, Sakai K, Yamamoto K, Hatanaka M, Tochikura T. Conversion of nitrite to nitrate by nitrite-resistant yeasts. Agricultural and Biological Chemistry, (1988); 52(8): 1999-2005.
- Zhang XY, Peng DC, Wan Q, Ju K, Wang BB, et al. Changing the nutrient source from ammonia to nitrate: Effects on heterotrophic bacterial growth in wastewater. Polish Journal of Environmental Studies, 2020; 29(2): 1473-1482.
- Romano N, Zeng C. Acute toxicity of sodium nitrate, potassium nitrate, and potassium chloride and their effects on the hemolymph composition and gill structure of early juvenile blue swimmer crabs (Portunus pelagicus Linnaeus, 1758) (Decapoda, Brachyura, Portunidae). Environmental Toxicology and Chemistry, (2007); 26(9): 1955-62.
- Prangnell DI, Samocha TM, Staresinic N. Water. In: Sustainable Biofloc Systems for Marine Shrimp (Samocha TM, ed.), (2019); 37. Academic Press, Elsevier Inc.
- Burford MA, Williams KC. The fate of nitrogenous waste from shrimp feeding. Aquaculture, (2001); 198(1-2): 79-93.
- Coelho RTI, Yasumaru FA, Passos MJACR, Gomes V, Lemos D. Energy budgets for juvenile Pacific white leg shrimp Litopenaeus vannamei fed different diets. Brazilian Journal of Oceanography, 2019; v67:e19243.
- Lin YC, Chen JC. Acute toxicity of nitrite on Litopenaeus vannamei (Boone) juveniles at different salinity levels. Aquaculture, (2003); 224(1-4): 193-201.
- Lin Y, Chen J. Acute toxicity of ammonia on Litopenaeus vannamei Boone juveniles at different salinity levels. Journal of Experimental Marine Biology and Ecology, (2001); 259(1): 109-119.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0














